4605
Denoising Magnetic Resonance Spectroscopy Signals with Stack Auto-encoder Networks
Jing Wang1, Bing Ji1, Yang Lei1, Tian Liu2, Hui Mao1, and Xiaofeng Yang1
1Emory University, Atlanta, GA, United States, 2Icahn School of Medicine at Mount Sinai, New York, NY, United States
1Emory University, Atlanta, GA, United States, 2Icahn School of Medicine at Mount Sinai, New York, NY, United States
Synopsis
Keywords: Signal Modeling, Spectroscopy
This abstract reports a novel and efficient self-supervised deep learning autoencoder network for denoising MRS data and improving signal-to-noise ratio (SNR) of spectra, which may enable rapid MRS data acquisition and improving its clinical workflow and applications.Introduction
Magnetic resonance spectroscopy (MRS) is a non-invasive imaging tool to identify biochemical and metabolic characteristics of tissue1. However, the concentrations of metabolites and biochemicals are often extremely low, e.g., in millimolar, for in vivo examinations. Thus, clinical applications of MRS are fairly limited due to low signal-to-noise ratios (SNR). To compensate for the low magnitude and SNR, typically a large sampling volume or voxel is used, or a high number of signal average (NSA) MRS is acquired to obtain sufficient SNR and interpretable quality spectra2. However, increasing voxel size reduces the spatial resolution and specificity, especially in heterogeneous tissues, while high NSA leads to prolonged scan time, as well as introduces time-related artifacts such as patient motion. One strategy to address this problem is to use denoising methods, such as smoothing filters and wavelet analysis which we reported previously3 to generate signal-only MR spectra with improved SNR. However, such methods require manually determined threshold values and possibly induce spectral linewidth broadening to compromise spectral resolutions. In this work, we report a fully automated deep-learning method to denoise MR spectra and improve SNR.Methods
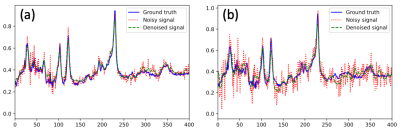
Denoising MRS data can be done with a stack auto-encoder (SAE) which is a self-supervised deep learning algorithm, by identifying and selecting noises, or noisy components, from noisy raw inputs, and then reconstructing “signal-only” spectra. Briefly, SAE encodes the noisy inputs in the compressing path and then recovers the denoised output in the expanding path. During the compressing path, the input information was encoded into hidden neurons and thus decomposed into various latent patterns, from which the noisy components were identified and removed. We used a spherical brain MRS phantom to obtain baseline high SNR or noisy MRS data for training the network, followed by testing our trained models using different sets of phantom data and MRS data collected from patients. For the phantom experiments, we used a single-voxel spectroscopy (SVS) PRESS sequence on a 3T Siemens scanner (Prisma) to record the MRS data in two locations of the phantom (front left (FL) and front right (FR)). At each location, we acquired high SNR MRS data (NSA=192) as raw inputs of MRS signals, while SVS MRS data from the human subjects were collected from the temporal lobe with an NSA of 192. For training and testing, the ground truth was considered to be the averaged MRS signal using 160 out of the 192 measurements. The noisy input was the averaged MRS signal using a smaller averaging number (NSA = 2 or 8) of the raw signals out of those 160 measurements. As a self-supervised SAE, the model tried to resemble the self-input with a much smoother output, so the training loss was defined as the mean squared error (MSE) between input and output signals. A denoising SAE was first trained on the phantom data collected at the FL location and then tested on the phantom FR location and the human temporal lobe. To overcome the curse of overfitting, we utilized a patch-based method to augment the datasets and then merged the denoised patches to recover smooth long signals. Two metrics of MSE and SNR were used to evaluate the model performance.Results
Our SAE MRS denoising model has shown clear improvement in the spectral quality and SNR after denoising, evidenced by decreased MSE and enhanced SNR in all tests. For inputs with high SNR (NSA= 8), the MSE decreased by 26.9% after denoising while SNR increased by 5.9% when the test was done with phantom data (e.g., FR); the MSE decreased by 45.6% and SNR increased by 12.8% for spectra collected from the temporal lobe of human subjects. For low SNR and noisy data (NSA = 2) inputs, the MSE decreased by 57.3% with SNR increased by 23.1% for the phantom data (FR). Similarly, MSE decreased by 66.5% and SNR increased by 32.6% for the data collected from the temporal lobe of human subjects. In all cases, the main high-magnitude peaks of brain metabolites were well preserved.Discussion
From the comparison of the noisy inputs (NSA of 2 and 8), we found that small NSA inputs demonstrated greater improvement. It could relate to the fact that low SNR data with NSA = 2 are much noisier than those collected with NSA=8. However, in principle both NSA = 2 and 8 signals embedded similar latent information. Thus, our SAEs were able to identify and select similar smooth patterns out of both types of noisy inputs. But for small SNR data (NSA = 2), though the major metabolite peaks were all preserved, some signals from the ultra-low concentration metabolites may be lost due to the intrinsic lack of information with limited inputs.Conclusion
We have shown that a deep-learning-based SAE denoising method can generate high-quality and high SNR MR spectra by persevering signals from key metabolites while removing noise identified from the noisy inputs. By applying the fully automated SAE denoiser, it is possible to utilize the otherwise noisy or low SNR spectroscopy data recorded with reduced acquisition time and size of sampling volume, thus improving MRS workflow and clinical adoptions of this molecular imaging method.Acknowledgements
No acknowledgement found.References
1. Dienel GA. Brain glucose metabolism: integration of energetics with function. Physiological reviews 2019;99(1):949-1045.
2. Shokrollahi P, Drake JM, Goldenberg AA. Signal-to-noise ratio evaluation of magnetic resonance images in the presence of an ultrasonic motor. Biomedical engineering online 2017;16(1):1-12.
3. Ji B, Hosseini Z, Wang L, Zhou L, Tu X, Mao H. Spectral Wavelet‐feature Analysis and Classification Assisted Denoising for enhancing magnetic resonance spectroscopy. NMR in Biomedicine 2021;34(6):e4497.
DOI: https://doi.org/10.58530/2023/4605