4603
Self-Supervised Model Fitting of VERDICT MRI in the Prostate1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Centre for Medical Imaging, University College London, London, United Kingdom, 3Molecular Diagnostics and Therapeutics Group, University College London, London, United Kingdom, 4Department of Urology, University College London Hospitals NHS Foundation Trust, London, United Kingdom
Synopsis
Keywords: Signal Modeling, Microstructure
Microstructure models are traditionally fitted via computationally expensive non-linear least squares. Recent model fitting techniques use supervised deep learning algorithms trained on synthetic datasets, however the training data distribution affects parameter estimates. Self-supervised learning can address this by extracting training labels directly from the input data. We introduce a self-supervised machine learning algorithm for fitting the VERDICT MRI model for prostate to diffusion-weighted MRI. On simulated data, our approach improves parameter estimation compared to non-linear least squares and supervised machine learning. We also reveal plausible tumour contrast on in-vivo prostate data.Introduction
Microstructure imaging combines bespoke diffusion-weighted MRI (DW-MRI) acquisitions with biophysical models to investigate sub-voxel tissue microstructure. These models are traditionally fitted using non-linear least squares (NLLS). Recent work has used supervised deep learning approaches to fit these models1-3. However, these are typically trained on large synthetic datasets, whose underlying distribution can introduce bias into fitted parameter estimates4. Self-supervised learning has the potential to address this issue by extracting training labels directly from the input data, but may also lead to higher variance5.Self-supervised model fitting has been used successfully to improve microstructural parameter estimation for two-compartment models5-7. This work is the first to trial this approach for a complex three-compartment model. We compare three model fitting strategies – NLLS, supervised learning, and our novel self-supervised learning approach – for the VERDICT-MRI model for prostate8-10, both in simulations and in in-vivo prostate. Results show that self-supervised fitting improves parameter estimation on simulated data compared to NLLS and supervised fitting, and yields maps with plausible tumour contrast on in-vivo scans.
Methods
Data AcquisitionData from one man with biopsy-confirmed clinically significant prostate cancer from the INNOVATE clinical trial11 was analysed in this study. VERDICT-MRI data was acquired on a 3T scanner (Achieva, Philips Healthcare, Best, Netherlands), with a pulsed-gradient spin echo sequence. Imaging parameters were: TR, 2482–3945 ms; TE 50–90 ms; field of view, 220 × 220 mm; section thickness, 5 mm; no intersection gap; acquisition matrix, 176 × 176; in-plane resolution, 1.25 × 1.25 mm; b-values, 90, 500, 1500, 2000, and 3000 s/mm2. Each b-value also included a b=0 image using the same TE as the corresponding diffusion weighted image, which was used to correct each volume for T2 effects. The total imaging time was 12 minutes12.
Simulated Dataset
10,000 DW-MRI signals were simulated to quantitatively assess the performance of the self-supervised fitting against NLLS and supervised fitting. The same acquisition parameters were used as in the patient dataset, and parameter values were randomly sampled (using a uniform distribution) from biophysically plausible parameter ranges: fIC, fEES: [0,1], R: [0,15]mm and dEES: [0.5,3] mm2/ms, with the constraint fIC + fEES + fVASC = 1. Rician noise was also added, with SNR = 50.
Biophysical Model
The VERDICT prostate model has three compartments that characterise diffusion in the vascular (VASC), extracellular-extravascular space (EES) and intracellular (IC) components in tumours. These are modelled as randomly-oriented sticks (AstroSticks), Gaussian free diffusion (Ball) and restriction in an impermeable sphere (Sphere) respectively8. We estimate four model parameters: fIC (IC volume fraction), fEES (EES volume fraction), cell radius R and diffusivity dEES13,14. The vascular volume fraction is calculated as fVASC = 1 – fIC – fEES. The DW-MRI signal for VERDICT is8:
S(b)/S0 = fVASCSVASC(dVASC,b) + fICSIC(dIC,R,b) + fEESSEES(dEES,b)
where b is the b-value and S0 is the b=0 signal intensity.
Parameter Estimation
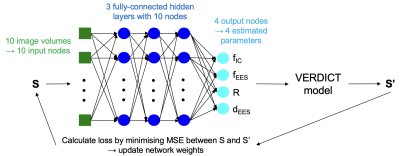
We consider three approaches: i) NLLS estimation, ii) supervised deep learning with a multilayer perceptron (MLP) consisting of three fully-connected hidden layers trained on 100,000 synthetic DW-MRI signals1,2 and iii) a novel self-supervised deep learning technique using a feed-forward, back propagation fully-connected neural network3. The network (shown in Figure 1) consists of an input layer and three fully-connected hidden layers (each with 10 neurons to match the number of diffusion-weighted acquisition volumes) and an output layer with four neurons, one for each of the parameters to be estimated.
Results
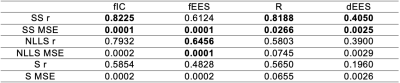
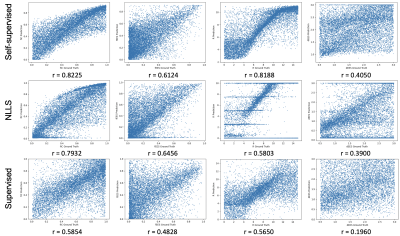
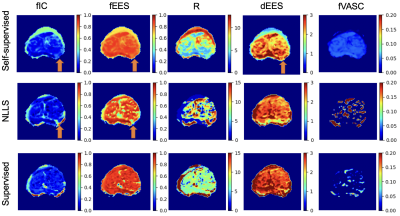
Table 1 shows mean squared error (MSE) values obtained between parameter estimates and ground truth values for all three fitting strategies on simulated data. We find that self-supervised fitting produces the lowest MSE across all four VERDICT parameters. Scatter plots of parameter estimates obtained using all three strategies against ground truth values are shown in Figure 2. Visually there is stronger correlation between estimates and ground truth using self-supervised fitting. This is supported by the Pearson correlation coefficients shown both on Figure 2 and in Table 1, which are higher for self-supervised fitting than for both other methods for fIC, R and dEES.All three fitting strategies are used to produce VERDICT parameter maps from in-vivo prostate data, which are shown in Figure 3. Self-supervised fitting produces maps with reasonable tumour contrast on in-vivo prostate data, but show increased homogeneity for fIC and fEES.
Discussion and Conclusion
This work presents a comparison between three DW-MRI model fitting strategies: NLLS, supervised deep learning via an MLP and a self-supervised FCNN. Self-supervised fitting demonstrates improved performance over supervised and NLLS fitting on simulated data, in terms of MSE and Pearson correlation coefficient. VERDICT parameter maps on in-vivo prostate data show reasonable tumour contrast, but are less detailed for certain parameters. This highlights the fact that although our self-supervised approach better estimates ground truth parameters in simulations, the ultimate goal is to reliably produce in-vivo maps with tumour contrast. In future, we will extensively tune our self-supervised network hyperparameters with the aim of revealing stronger tumour contrast in real MRI data.Future work will quantitatively compare the fitting strategies in more detail by considering bias and variance separately. We will also analyse their performance on a larger patient cohort in delineating different tissue types, and explore the generalisability of self-supervised learning to unseen data.
Acknowledgements
This work is supported by the EPSRC-funded UCL Centre for Doctoral Training in Intelligent, Integrated Imaging in Healthcare (i4health) (EP/S021930/1) and the Department of Health’s NIHR-funded Biomedical Research Centre at University College London Hospitals. This work is also funded by EPSRC grant numbers EP/N021967/1, EP/R006032/1, EP/V034537/1; and by Prostate Cancer UK, Targeted Call 2014, Translational Research St.2, grant number PG14-018-TR2. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.References
| |||||||||||||||||||||||||||||||
Figures