4599
Simultaneous numerical modeling of 13C-MRI enzyme kinetics, diffusion effects, and RF refocusing1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Physical Sciences Platform, Sunnybrook Research Institute, Toronto, ON, Canada
Synopsis
Keywords: Signal Modeling, Hyperpolarized MR (Non-Gas), 13C Signal Dynamics, 13C MRI, Enzyme Kinetics, Reaction Kinetics, Metabolism
In this study, we used a particle-based Brownian dynamics simulator with concurrent MR modeling to characterize diffusion-related effects on 13C signal dynamics. We conducted a spectrophotometric lactate dehydrogenase (LDH) activity assay with and without the presence of glass beads to measure the influence of diffusion constraints and spatial compartmentalization on the reaction kinetics of LDH-mediated conversion of pyruvate to lactate. We also investigated the relationship between diffusion and RF refocusing in a typical fast GRE sequence by comparing simulated signal intensities for RF spoiled and non-spoiled GRE sequences applied under varying diffusion conditions.Introduction
Hyperpolarized (HP) carbon-13 MRI is a powerful tool for metabolic imaging, but interpretation of in vivo results is challenging due to the complexity of 13C signal dynamics, especially the variety of factors which affect metabolic reaction kinetics.1,2 Reaction rates involving 13C metabolites are influenced by diffusion, transport, and uptake processes, which vary regionally and locally due to differences in cellular microstructure and metabolite compartmentalization.1–5 Characterizing the interplay between microstructural and biochemical influences on reaction rates is made even more difficult by the non-recoverable nature of hyperpolarized signal, which limits HP 13C MRI scans to a relatively narrow time window. Gradient-echo (GRE) pulse sequences are useful for applications where rapid acquisition is important, such as in 3D imaging;6 however, if the repetition time of a GRE sequence is short relative to relaxation time, then successive acquisitions are affected by transverse coherences.6,7 Fast GRE sequences without RF spoiling are often used for HP 13C data acquisition,8–11 resulting in a contribution from refocused transverse magnetization that is diffusion-weighted and may contribute significantly to the metabolite signal.Methods
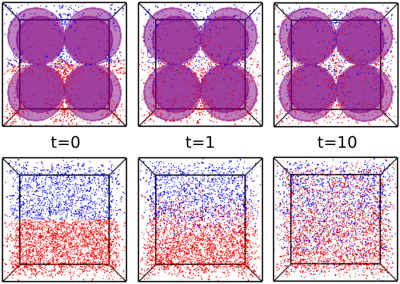
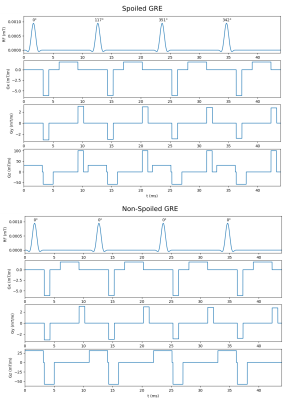
To assess reaction kinetics of LDH-mediated conversion of pyruvate to lactate, a standard LDH activity assay protocol was used with a range of LDH concentrations.13 For each assay, 0.5 mL of sodium pyruvate in phosphate buffer (pH 7.3) was added to a polystyrol cuvette, followed by 0.5 mL of reduced nicotinamide adenine dinucleotide (NADH), for final concentrations of 2 mM and 1 mM respectively. Lastly, LDH was added to concentrations from 0.8-2.8 units/mL, and decrease in absorbance at 340 nm was monitored at 1 s intervals to determine the maximum reaction rate (Vmax) for each run. Assays at each concentration of LDH were conducted twice, once with and once without the presence of glass beads (5 mm diameter) as a model of structural constraints on diffusion. This system was reproduced in Smoldyn by generating a cubic volume with 10 mm isotropic length containing either 0 or 8 spherical excluded volumes with 5mm diameter. The lower half of this volume was populated with 1000 pyruvate particles and the upper half with 500-2000 LDH particles (shown in Figure 1), with a bidirectional reaction on contact between particles allowing conversion between pyruvate and LDH. MR spectroscopic measurements were taken by applying a 90° pulse on pyruvate at 1 s intervals and reading out the total transverse magnetization.For RF refocusing experiments, simulated images were generated in our Smoldyn MR module using two versions of a 64x64 gradient echo sequence, one with RF spoiling via a standard phase cycling scheme14 and crusher gradients applied in the z-direction, and the other with no spoiling (partially shown in Figure 3). In each simulated scan, 1000 particles were deposited into a rectangular central volume (dimensions 10 mm x 10 mm x 1 mm) and set to diffuse freely with varying diffusion coefficients. Signal intensity was calculated from the resulting images by taking the summed voxel intensity from the region of interest. All simulations were conducted on 1-4 cores of an AMD Ryzen 9 5900X 12-Core Processor operating at 4.80 GHz.
Results and Discussion
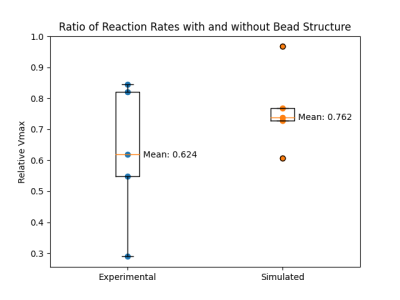
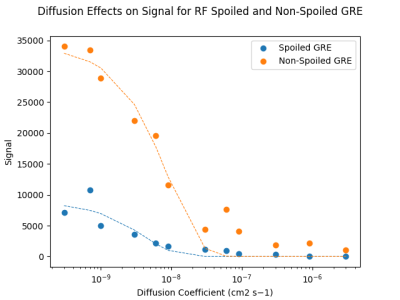
We previously reported application of a Smoldyn-based MR model to monocarboxylate reaction kinetics, with close agreement between simulated and in vitro measured results.15 By introducing a structural constraint on diffusion through addition of glass beads (Figure 1), a consistent reduction in the reaction rate of LDH-mediated pyruvate to lactate conversion was observed (Figure 2), with a similar magnitude in vitro (mean Vmax ratio of 0.624) and in silico (mean Vmax ratio of 0.762). The higher variability in the in vitro results is likely primarily due to differences in the bead arrangement, which affects both the model structure and the effective path length of the spectrophotometric measurement. Future work will build on these results by using MR phantoms with similar internal structure to model MR signals expected from different microstructural conditions.In addition to modeling how changes to diffusion conditions influence reaction kinetics, we investigated how such changes to diffusion rates affect MR signal intensity in GRE sequences with and without the use of RF spoiling and crusher gradients to eliminate residual magnetization between successive excitations. As shown in Figure 4, signal intensities from images acquired with the spoiled GRE sequence were consistently lower than from the non-spoiled sequence, reflecting the loss of signal contribution from residual magnetization. Signal intensities for both spoiled and non-spoiled sequences generally declined sharply with higher diffusion coefficients; variability in this trend for some data points was likely a result of the lower particle count (1000 per simulation) used in this experiment.
Conclusions
An in silico model using particle-based Brownian dynamics and concurrent MR data acquisition and simulation was compared with spectrophotometric lactate dehydrogenase (LDH) activity measurements under two different conditions (with and without glass beads) and was in rough agreement. The in silico model was also shown to reproduce complex MR signal mechanisms such as the RF refocusing that occurs in fast gradient-echo sequences. The in silico model resulting from this work will be useful for designing and predicting the outcome of future in vivo experiments to understand metabolic compartmentalization in tissue.Acknowledgements
The authors are grateful for funding from Canadian Institutes for Health Research grant CIHR PJT152928 and Natural Sciences and Engineering Research Council grant RGPIN-2016-05566.
References
1. Hurd, R. E., Yen, Y.-F., Chen, A. & Ardenkjaer‐Larsen, J. H. Hyperpolarized 13C metabolic imaging using dissolution dynamic nuclear polarization. J. Magn. Reson. Imaging 36, 1314–1328 (2012).
2. Ross, B. D., Bhattacharya, P., Wagner, S., Tran, T. & Sailasuta, N. Hyperpolarized MR Imaging: Neurologic Applications of Hyperpolarized Metabolism. Am. J. Neuroradiol. 31, 24–33 (2010).
3. Søgaard, L. V., Schilling, F., Janich, M. A., Menzel, M. I. & Ardenkjær-Larsen, J. H. In vivo measurement of apparent diffusion coefficients of hyperpolarized 13C-labeled metabolites. NMR Biomed. 27, 561–569 (2014).
4. Koelsch, B. L. et al. Rapid in vivo apparent diffusion coefficient mapping of hyperpolarized 13C metabolites. Magn. Reson. Med. 74, 622–633 (2015).
5. Zhu, X., Gordon, J. W., Bok, R. A., Kurhanewicz, J. & Larson, P. E. Z. Dynamic Diffusion Weighted Hyperpolarized 13C Imaging Based on a Slice-Selective Double Spin Echo Sequence for Measurements of Cellular Transport. Magn. Reson. Med. 81, 2001–2010 (2019).
6. Hargreaves, B. Rapid Gradient-Echo Imaging. J. Magn. Reson. Imaging JMRI 36, 1300–1313 (2012).
7. Bernstein, M. A., King, K. F. & Zhou, X. J. Handbook of MRI Pulse Sequences. (Elsevier, 2004).
8. Cunningham, C. H. et al. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. J. Magn. Reson. 187, 357–362 (2007).
9. Cunningham, C. H. et al. Pulse Sequence for Dynamic Volumetric Imaging of Hyperpolarized Metabolic Products. J. Magn. Reson. San Diego Calif 1997 193, 139–146 (2008).
10. Geraghty, B. J., Lau, J. Y. C., Chen, A. P. & Cunningham, C. H. Dual-Echo EPI sequence for integrated distortion correction in 3D time-resolved hyperpolarized 13C MRI. Magn. Reson. Med. 79, 643–653 (2018).
11. Lee, C. Y. et al. Lactate topography of the human brain using hyperpolarized 13C-MRI. NeuroImage 204, 116202 (2020).
12. Andrews, S. S. Smoldyn: particle-based simulation with rule-based modeling, improved molecular interaction and a library interface. Bioinformatics 33, 710–717 (2017).
13. Tegge, G. Bergmeyer, H. U. (Editor-in-Chief): Methods of Enzymatic Analysis (Methoden der enzymatischen Analyse), 3rd Edition. Editors: J. Bergmeyer und Marianne Graßl. Volume III, Enzymes 1: Oxidoreductases, Transferases. Verlag Chemie, Weinheim – Deerfield Beach – Basel 1983. XXVI, 605 p., with 18 figs. and 43 tables. Hardcover-cloth DM 224,– (subscription price, when all 10 volumes are ordered) individual volume price DM 258,–. Starch - Stärke 37, 106–107 (1985).
14. Zur, Y., Wood, M. L. & Neuringer, L. J. Spoiling of transverse magnetization in steady-state sequences. Magn. Reson. Med. 21, 251–263 (1991).
15. (ISMRM 2022) Novel particle-based spatial stochastic Bloch simulation applied to LDH-mediated pyruvate conversion. https://archive.ismrm.org/2022/1425.html.
Figures