4552
Development of a MR-compatible bioreactor for 3D cell structure metabolic analysis in real-time for a 60 MHz benchtop NMR spectrometer.1Institute for Bioengineering of Catalonia, Barcelona, Spain
Synopsis
Keywords: New Devices, Cancer, Metabolism
Bioreactor platforms have been used in HP-NMR applications to analyze cellular metabolism in real-time, however they require complex set-ups and automated controlled systems, limiting their use. We present a system that overcomes these limitations consisting of a 3D printed platform, an NMR compatible 3D cell model and a recirculation circuit for single-shot and longitudinal hyperpolarization-enhanced NMR experiments. The platform presents no signal interference, a media system for biological support, and a straightforward and simplified manufacturing and assembly process. As a proof-of-concept, we present preliminary metabolic data of a 3D HeLa cell model analyzed with our bioreactor.
Introduction
In recent years, dissolution Dynamic Nuclear Polarization (dDNP) has been used to increase the sensitivity of NMR by over 50.000-fold compared to conventional NMR acquisitions2. This enhancement enables tracking metabolic conversions and studying cellular metabolism in real-time while causing no damage to the sample.Bioreactor platforms have been used to probe cell metabolism by DNP-NMR. Usually, these bioreactors use complex set-ups and automated controlled systems3,4, which results in platforms difficult to prepare and operate due to special fabrication requirements. Here we present an alternative low-cost bioreactor - a 3D printed design to simplify its in-house fabrication and assembly - for improved accessibility in research and clinical settings. This platform has been designed to carry out DNP-NMR analyses of 3D cell models and has been tested on a tissue engineered model of cervical cancer developed in our laboratory.
Methods
NMR-compatible bioreactor design and fabrication: The design of the bioreactor parts (Figure 1) was done using computer-aided design (CAD) software. The pieces were printed using a Microlay Versus 385nm 3D Printer (Microlay SLA Systems, ES; XY resolution of 65 μm, Z resolution of 10 μm) with a biocompatible resin (Freeprint® Ortho, 03988, DETAX, DE). The printed pieces were assembled into functional bioreactors with capillary tubes using heat shrink tubes as connectors and insulators.NMR-compatible bioreactor validation: To assess signal interference, the 1H spectral signal of water was acquired with and without the bioreactor placed inside a 5-mm NMR tube using a 1.4 T Benchtop Spectrometer (Pulsar, Oxford Instruments).
3D HeLa model and viability assay under flow conditions: One million HeLa cells (human cervical adenocarcinoma cells) were seeded in 1% CMC cryogels5 for metabolic analysis. The cell-laden scaffold was placed into a 5mm NMR tube with the bioreactor connected to the fluidic circuit through a peristaltic pump for media recirculation and gas exchange. After 20h, cell viability was calculated through metabolic activity by alamarBlueTM assay (ThermoFischer Scientific, DAL1025). These results were compared to the ones obtained after analyzing constructs kept in static conditions both in a well plate and in an NMR tube. The biochemical assays were complemented with staining and confocal microscopy images from a LSM 800 Leica (Carl Zeiss) scanning laser confocal microscope operating with ZEN 2.3 (blue edition) imaging software.
Results and Discussion
Our MR-compatible bioreactor design is ideal for an easy and non-specialized fabrication, assembly and use. The design allows for media recirculation, fast dDNP solution injection, and precise positioning of the 3D cell-laden scaffold in the detection area of the RF coil. Figure 1 shows the individual components and their assembly. The platform is NMR compatible (Fig. 2) with a FWHM of 2.0 Hz for the bioreactor and a FWHM of 2.5 Hz for bioreactor and scaffold, having minor effect on the line shape. The 3D cell model has been specially designed to overcome some of the challenges that DNP-NMR presents, namely fast diffusion of the dDNP substrate and low signal susceptibility from the biomaterial. The cryogel scaffolds used here allow the cells to self-arrange and distribute inside the biomaterial while providing structure, a closer spatial distribution to that found in the native tissue. Confocal images show (Fig. 3, D) a homogeneous spatial distribution of the cells inside the 1% CMC scaffold, moreover, no cell death can be observed throughout sample pool. Additionally, in-flow experiments enabled by this bioreactor show a 10% increase on the metabolic signal when compared to the models maintained in culture conditions, and 85% increase when compared to those models maintained in the NMR tube in static conditions (Fig. 3, E). These results show that cells remain more metabolically active inside the NMR tube with a media recirculation system facilitated by our bioreactor.Conclusions
We developed an easy and robust MR-compatible bioreactor for single-shot and longitudinal NMR experiments where a fluidic system keeps 3D cell models viable and metabolically active for NMR experiments without hampering signal acquisition. Combining tissue engineered models and the proposed bioreactor, we can study cell metabolism in a more physiologically accurate environment that can be maintained for reproducible experiments at several time points.Acknowledgements
This work is part of a project that has received funding from the Junior Leader Postdoctoral Fellowship Programme from “la Caixa” Banking Foundation (LCF/BQ/ PI18/11630020), MCIN/AEI/10.13039/501100011033 (Ref. PID2020-117859RA-I00), the European Union’s Horizon 2020 research and innovation program (GA-863037), the BIST – “la Caixa” initiative in Chemical Biology (CHEMBIO), the FI Fellowship Programme from AGAUR (Ref. 2021 FI_B_01039), Grant RYC2020-029099-I funded by MCIN/AEI/ 10.13039/501100011033 and by “ESF Investing in your future”, Marie Skłodowska-Curie grant (Ref.101063517).References
1. Kayvan R. Keshari, John Kurhanewicz, Rex E. Jeffries, David M. Wilson BJ, Dewar, Mark Van Criekinge, Matthew Zierhut, Daniel B. Vigneron and JM, Macdonald. Hyperpolarized 13C Spectroscopy and an NMR-Compatible Bioreactor System for the Investigation of Real-Time Cellular Metabolism. Magn Reson Med. 2010;63(2):322-329. doi:10.1002/mrm.22225.Hyperpolarized
2. Yoshihara HAI, Can E, Karlsson M, Lerche MH, Schwitter J, Comment A. High-field dissolution dynamic nuclear polarization of [1-13C]pyruvic acid. Physical Chemistry Chemical Physics. 2016;18(18):12409-12413. doi:10.1039/c6cp00589f
3. Sriram R, Nguyen J, Santos JDL, et al. Molecular detection of inflammation in cell models using hyperpolarized 13 C-pyruvate. Theranostics. 2018;8(12):3400-3407. doi:10.7150/thno.24322
4. Mancuso A, Beardsley NJ, Wehrli S, Pickup S, Matschinsky FM, Glickson JD. Real-time detection of 13C NMR labeling kinetics in perfused EMT6 mouse mammary tumor cells and βHC9 mouse insulinomas. Biotechnol Bioeng. 2004;87(7):835-848. doi:10.1002/bit.20191
5. Velasco-Mallorquí F, Rodríguez-Comas J, Ramón-Azcón J. Cellulose-based scaffolds enhance pseudoislets formation and functionality. Biofabrication. 2021;13(3). doi:10.1088/1758-5090/ac00c3Figures
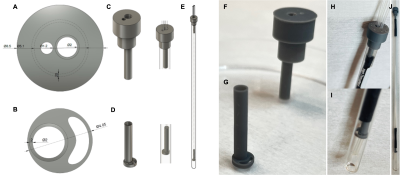
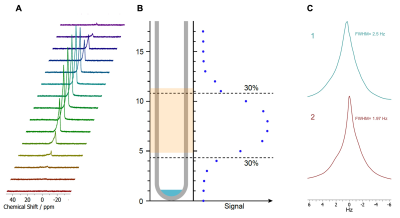

Figure 3. Characterization of 3D HeLa cell model. (A) Structure of the cryogel through brightfield optical microscopy. (B) Structure and dimensions of the cryogel at the naked eye. (C) Structure of the dehydrated cryogel through scanning electron microscopy. (D) HeLa cell clusters self-assembled in the cryogel after seeding. (E) Metabolic viability of 3D samples after 20h without flow in the NMR tube, in static conditions in a well plate and under flow conditions in the bioreactor: *p<0.03.