4533
Can Zero Echo Time (ZTE) Magnetic Resonance Imaging Sequences Detect Signal from Collagen Backbone Protons?1Radiology, University of California San Diego, San Diego, CA, United States, 2Radiology, GE Healthcare, San Diego, CA, United States, 3VA San Diego Healthcare System, San Diego, CA, United States, 4Bioengineering, University of California San Diego, San Diego, CA, United States
Synopsis
Keywords: Bone, Bone
Ultrashort echo time (UTE) and zero echo time (ZTE) sequences have been extensively investigated for imaging of short T2 species. The ZTE sequence has a shorter effective TE than UTE and may be superior in imaging ultrashort T2 species. This study investigated whether ZTE could directly image collagen backbone protons in bovine cortical bone and human patellar tendon samples after D2O exchange and freeze-drying. Our experimental results demonstrate that collagen backbone protons are "invisible" with ZTE, which may not be able to directly image species with T2s of ~10 µs.Introduction
The detection of collagen changes is clinically and scientifically important because it is the most abundant protein in the human body (1–3). Collagen backbone protons have extremely short T2s or T2*s of the order of ~10 microseconds (4–6). Advanced MRI techniques, such as ultrashort echo time (UTE) and zero echo time (ZTE) sequences, have facilitated short-T2 imaging (2,5,6). Several studies have claimed that ZTE sequences can detect signals from semisolids more efficiently than UTE sequences (7,8). This study investigates the feasibility of ZTE sequences in detecting signals from collagen backbone protons via D2O exchange and freeze-dry studies of cortical bone and patellar tendon specimens at 3T.Method
Ten bovine cortical bone samples (~30×10×5mm3) and one human patellar tendon sample (8cm in length) were prepared for this study. All samples were imaged with three-dimensional ZTE and UTE sequences on a 3T clinical scanner (GE Healthcare Technologies, Milwaukee, MI, USA). Figure 1 shows the ZTE and UTE sequence diagrams. ZTE employed a non-selective rectangular RF pulse with a short duration (duration=8μs) for excitation, followed by 3D center-out radial sampling (8). The 3D UTE Cones sequence utilized a short rectangular pulse (duration=32μs) for non-selective excitation, followed by 3D radial sampling with conical view ordering (9). A 4-channel wrist coil was used for signal reception. For the ZTE sequence, the following parameters were used: TR=2.1ms, flip angle=4°, bandwidth=62.5kHz, field of view=4cm, 52 slices, slice thickness=3mm, and reconstruction matrix=192×192×16 for cortical bone and 256×256×16 for patellar tendon. Similar imaging parameters were used for the 3D UTE sequence except for a longer TR of 10ms and a higher flip angle of 10°.Results
Figures 2 and 3 show 3D ZTE and UTE images of the bovine cortical bone sample and human Patellar tendon sample, before(A, B, respectively) and after (C, D, respectively) D2O-H2O exchange followed by freeze-drying. A high signal was detected for all samples before the procedures. However, after the procedures, pure noise was observed for all bone samples, and only a thin bright line was observed in the margins of the patellar tendon sample. The thin bright line was later found from fat and showed typical fat/water in-phase and out-phase behaviors. The D2O exchange and freeze-drying procedures removed all water components, leaving collagen backbone protons being selectively imaged with the ZTE and 3D-UTE sequences. The pure signal void suggests neither UTE nor ZTE can directly detect signals from collagen backbone protons in cortical bone and the patellar tendon.Discussion and conclusion
This is the first study investigating the feasibility of directly imaging collagen protons using the 3D ZTE technique. Ma et al. reported that 2D and 3D UTE sequences could not directly image the collagen matrix(4). Wu et al.(7) reported that proton water- and fat-suppressed projection MR imaging (WASPI) could directly image the solid matrix Siu et al.(10) reported that UTE sequences could detect signals from collagen protons at 7T (10) The contradictory results highlight the importance of this study - whether collagen protons are "visible" with ZTE. The strong ZTE signal from bone and tendon samples before D2O-exchange and freeze-drying but pure noise after demonstrate that collagen backbone protons are “invisible” with the ZTE sequence. One way to indirectly image collagen backbone protons is UTE magnetization transfer (UTE-MT) imaging and signal modeling. Recent UTE-MT studies suggest that collagen backbone protons have(6,11) extremely short T2s of 6.4-15.6 µs(6,12). The T2 values are largely consistent with macromolecular proton T2s reported in the literature(4,13,14). Our experimental results suggest that ZTE cannot directly image species with T2s of ~10 µs.Acknowledgements
The authors acknowledge grant support from the National Institutes of Health (R01AR062581, R01AR068987, R01AR075825, R01AR079484, R01AR078877, RF1AG075717, and R21AR075851), VA Clinical Science and Rehabilitation Research and Development Services (Merit Awards I01CX001388, I01CX002211, and I01RX002604), and GE Healthcare.References
1. Scholarlycommons S, Li C. Magnetic Resonance Imaging of Short-T2 Tissues with Magnetic Resonance Imaging of Short-T2 Tissues with Applications for Quantifying Cortical Bone Water and Myelin Applications for Quantifying Cortical Bone Water and Myelin [Internet]. Available from: https://repository.upenn.edu/edissertations/1350
2. Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging. J Comput Assist Tomogr [Internet]. 2003 [cited 2022 Nov 4];27(6):825–46. Available from: https://pubmed.ncbi.nlm.nih.gov/14600447/
3. di Lullo GA, Sweeney SM, Körkkö J, Ala-Kokko L, San Antonio JD. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J Biol Chem [Internet]. 2002 Feb 8 [cited 2022 Nov 4];277(6):4223–31. Available from: https://pubmed.ncbi.nlm.nih.gov/11704682/
4. Henkelman RM, Huang X, Xiang Q ‐S, Stanisz GJ, Swanson SD, Bronskill MJ. Quantitative interpretation of magnetization transfer. Magn Reson Med [Internet]. 1993 [cited 2022 Nov 8];29(6):759–66. Available from: https://pubmed.ncbi.nlm.nih.gov/8350718/
5. Chen Y, Li L, Le N, Chang EY, Huang W, Ma YJ. On the fat saturation effect in quantitative ultrashort TE MR imaging. Magn Reson Med. 2022 May 1;87(5):2388–97.
6. Hodgson RJ, Evans R, Wright P, Grainger AJ, O’connor PJ, Helliwell P, et al. Quantitative magnetization transfer ultrashort echo time imaging of the Achilles tendon. Magn Reson Med [Internet]. 2011 May 1 [cited 2022 Nov 7];65(5):1372–6. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/mrm.22715
7. Wu Y, Ackerman JL, Chesler DA, Graham L, Wang Y, Glimcher MJ. Density of organic matrix of native mineralized bone measured by water- and fat-suppressed proton projection MRI. Magn Reson Med. 2003 Jul 1;50(1):59–68.
8. Weiger M, Pruessmann KP. MRI with Zero Echo Time. In: Encyclopedia of Magnetic Resonance. John Wiley & Sons, Ltd; 2012.
9. Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med [Internet]. 2006 [cited 2022 Nov 5];55(3):575–82. Available from: https://pubmed.ncbi.nlm.nih.gov/16450366/
10. Siu AG, Ramadeen A, Hu X, Morikawa L, Zhang L, Lau JYC, et al. Characterization of the ultrashort-TE (UTE) MR collagen signal. NMR Biomed [Internet]. 2015 Oct 1 [cited 2022 Nov 4];28(10):1236–44. Available from: https://pubmed.ncbi.nlm.nih.gov/26268158/
11. Chang EY, Bae WC, Shao H, Biswas R, Li S, Chen J, et al. Ultrashort Echo Time Magnetization Transfer (UTE-MT) Imaging of Cortical Bone. NMR Biomed [Internet]. 2015 Jul 1 [cited 2022 Nov 5];28(7):873. Available from: /pmc/articles/PMC4652942/
12. Ma YJ, Chang EY, Carl M, Du J. Quantitative magnetization transfer ultrashort echo time imaging using a time-efficient 3D multispoke Cones sequence. Magn Reson Med [Internet]. 2018 Feb 1 [cited 2022 Nov 8];79(2):692–700. Available from: https://pubmed.ncbi.nlm.nih.gov/28470838/
13. Ramani A, Dalton C, Miller DH, Tofts PS, Barker GJ. Precise estimate of fundamental in-vivo MT parameters in human brain in clinically feasible times. Magn Reson Imaging. 2002 Dec 1;20(10):721–31.
14. Sled JG, Bruce Pike G. Quantitative imaging of magnetization transfer exchange and relaxation properties in vivo using MRI. Magn Reson Med [Internet]. 2001 Nov 1 [cited 2022 Nov 8];46(5):923–31. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/mrm.1278
Figures
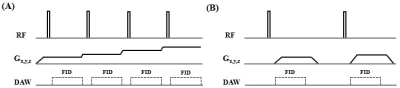
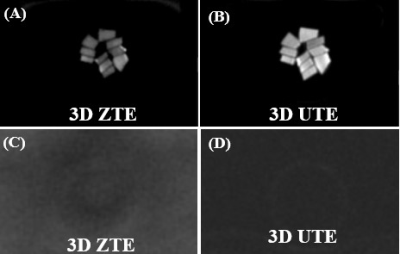
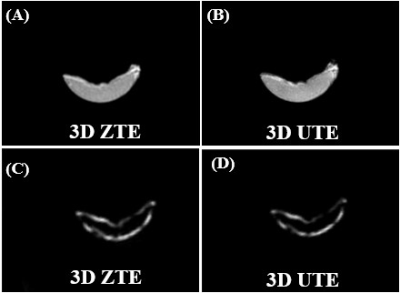
Figure 3. A cadaveric human patellar tendon was imaged with 3D ZTE (A) and 3D UTE (B) at 3T, as well as 3D ZTE (C) and 3D UTE (D) imaging of the same patellar specimen after 2 days D2O exchange followed by freeze–drying for over 40 hours. The patellar tendon sample was visible before freeze–drying (A, B), but invisible after freeze–drying (C, D). The thin bright line was from fat and showed typical fat/water in-phase and out-phase behaviors.