4528
Intelligent volume rendering of ZTE MR Bone images
Vineet Mehta1, Deepthi Sundaran1, Jignesh Dholakia1, Maggie Fung2, and Michael Carl3
1GE Healthcare, Bengaluru, India, 2GE Healthcare, New York City, NY, United States, 3GE Healthcare, San Diego, CA, United States
1GE Healthcare, Bengaluru, India, 2GE Healthcare, New York City, NY, United States, 3GE Healthcare, San Diego, CA, United States
Synopsis
Keywords: Bone, Visualization, Volume Rendering
This study presents a new Deep-Learning (DL) based processing pipeline to enable one-click volume rendering of MR Bone Zero TE (ZTE) images. It comprises of DL based background subtraction followed by histogram adjustment. A comparison with existing algorithms demonstrates that the proposed method prevails in accuracy of background subtraction (DICE = 0.97) and eliminates the tedious, error-prone manual steps of segmentation and histogram adjustment required to prepare the data for usable volume rendering. It can also be used universally across anatomies as opposed to other methods requiring anatomy-specific tuning.Introduction
MR Bone Zero echo time (oZTEo) imaging is a relatively recent development in MR technology that produces images similar to CT or radiography. The major application of ZTE is musculoskeletal imaging which can obviate the need for an additional modality that uses ionizing radiations like CT or X-ray. However, 3D volume rendering using ZTE is currently time-consuming and require postprocessing software that features manual contouring/segmentation1. The major reason for this tiresome process is the high intensity of the background (unlike CT) which is similar to the intensity of the bone pixels and the variable dynamic range. To this end, we propose an intelligent volume rendering (as shown in Figure 1) of ZTE images which consists of two steps: - 1) automated background subtraction and 2) histogram adjustment to handle dynamic range.Background subtraction has a lot of applications in computer vision related tasks and is often formulated as two class segmentation problem namely background and foreground. While the traditional methods of segmentation include thresholding, region-based methods, clustering etc.; the recent state-of-the-art methods include deep learning and convolutional neural networks which have also widely been adopted for medical image segmentation2,4,5.
Methods
U-net SegmentationFor background subtraction, the standard U-Net3 model with 4 encoding and 4 decoding blocks was trained. For pre-processing, images were resized to 256x256 and rescaled using min-max normalization. Due to the skewed anatomy distribution, extensive data augmentation strategies such as crop, flip, rotation and multiplanar reformats were used to improve the model robustness. The model was trained with cross entropy loss and Adam optimizer.
Data
The dataset consisting of 86 DICOM series with variations in magnetic field strength, orientation and anatomy were used for experiments. The segmentation masks for 62 series datasets were manually labelled by clinical experts while the rest of them were annotated by traditional image segmentation methods, which together constituted the ground truth. Air in the background & lungs are considered background in the manual labels. One exception is that for head/neck cases, air inside the skull or airways are not considered background in the manual labels in this experiment because of lack of accurate and consistent methods to segment it out manually. In order to ensure an even distribution of anatomy and orientation among training, test and validation sets, the datasets were grouped by the nearest canonical orientation and anatomy coverage. In total, the dataset consisted of 9310 instances / images (train - 54%, validation - 15%, test - 31%) across 11 anatomical categories as shown in Figure 2.
Volume Rendering
Once the background is effectively suppressed, as a next step, the histogram of the image was adjusted such that bone and tissue falls into a specific range for all ZTE images as shown in Figure 3. The main peak of the histogram was shifted to a value of 1000, which corresponds to the Hounsfield unit for bone. In addition, some minor additional hole filling and skin erosion were performed after the DL inferencing to improve visualisation.
Results and Discussion
For background subtraction, the proposed deep learning-based model was tested across 26 DICOM series and the results were compared against the ground truths and the traditional image segmentation method as shown in Figure 4. The results and observations can be summarized as: -- The proposed model achieved an average DICE score of 0.97. As seen in Figure 4, the DL model performed much better than the traditional method. It was found to be more robust to variations in anatomy especially in the cases which involves metal implants (Figure 4.b) and the lungs along with airway (Figure 4.c).
- The model is found to miss bright bone area nearby boundary in some cases (Figure 4.e) which could be due to less data for these cases, or the need to incorporate boundary awareness in the deep learning-based method. Nevertheless, these losses can be easily recovered through post processing using morphological operations.
Conclusion
The processing pipeline comprising of DL based background subtraction followed by histogram adjustment is described to achieve one-click volume rendering of ZTE images. The DL based method for background subtraction performed better than the traditional method across a wide range of anatomies and acquisition planes. The images generated from the pipeline reduced the time required for generating good volume rendered images.Acknowledgements
We would like to thank GE Healthcare for providing us with the necessary resources for this work. Also, a special thanks to Ananthakrishna Madhyastha for his continuous encouragement and support.References
- Aydıngöz Ü, Yıldız AE, Ergen FB. Zero Echo Time Musculoskeletal MRI: Technique, Optimization, Applications, and Pitfalls. RadioGraphics. 2022 Sep;42(5):1398-414.
- Lei T, Wang R, Wan Y, Du X, Meng H, Nandi AK. Medical Image Segmentation Using Deep Learning: A Survey.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. InInternational Conference on Medical image computing and computer-assisted intervention 2015 Oct 5 (pp. 234-241). Springer, Cham.
- Agarwal, A., Goel, M., & Dholakia, J. 3-plane Localizer-aided Background Removal in Magnetic Resonance (MR) Images Using Deep Learning.
- Sundaran D, Kulkarni D, Dholakia J. Optimal Windowing of MR Images using Deep Learning: An Enabler for Enhanced Visualization. arXiv preprint arXiv:1908.00822. 2019 Aug 2
Figures
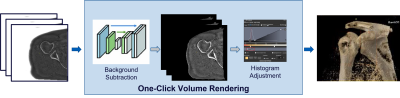
Figure 1: The proposed one-click volume rendering consisting of Deep Learning based background subtraction followed by histogram adjustment.
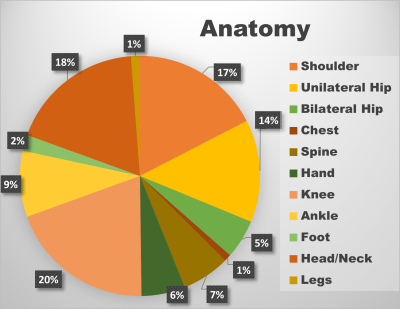
Figure 2: Data distribution across anatomy.
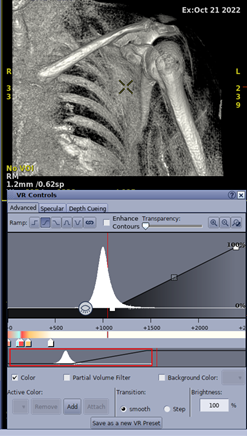
Figure 3: Histogram Adjustment.
Figure 4: Comparison of the Unet predictions against the traditional method output and the ground truths.
DOI: https://doi.org/10.58530/2023/4528