4519
Reduced input combination study for the Simultaneous Multi-Tissue Segmentation and Multi-Parameter Quantification Network (MSMQ-Net) of Knee1Department of Radiology, University of California, San Diego, San Diego, CA, United States, 2Department of Neurosciences, University of California, San Diego, San Diego, CA, United States, 3Radiology Service, Veterans Affairs San Diego Healthcare System, San Diego, CA, United States
Synopsis
Keywords: Osteoarthritis, Quantitative Imaging
To accelerate ultrashort echo time (UTE) based multi-parameter quantitative MRI (qMRI), we performed comparison studies on the input MRI image numbers and the effects on the prediction quality of the MSMQ-Net. The MSMQ-net was modified and trained accordingly with different combinations of inputs. Both image similarity and regional analysis were evaluated. The results demonstrate that 90% accuracy can be achieved for both UTE-T1 and UTE-T1rho mapping when the scan time is reduced by 75%.Introduction
Convolutional Neural Network(CNN) based deep learning has been widely used in the MRI area, such as segmentation, registration, reconstruction, and quantitative mapping1. Based on the encoder-decoder structure of the CNN, we proposed Multi-Tissue Segmentation and Multi-Parameter Quantification network (MSMQ-Net), which can obtain the segmentation and qMRI mapping simultaneously to accelerate the pipeline for the region-based qMRI analysis2,3. It is critically important to generate robust multi-parameter mapping with reduced input images to shorten the total scan time. In this study, we performed comparison studies on the input ultrashort echo time (UTE) MRI image numbers and the effects on the prediction quality of the MSMQ-Net.Method
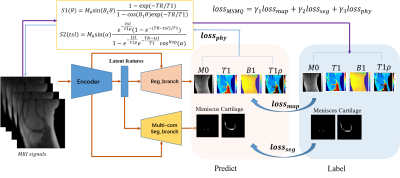
As shown in Figure 1, MSMQ-Net is based on a U-Net style DCNN with two branches for outputs, namely Reg_branch and Seg_branch. Along the encoder path, downsamplings are applied to enable subsequent feature extractions at coarser scales. Latent features are shared for both up-sampling branches, and up-samplings are conducted to support subsequent feature extractions at finer scales. Between convolutional blocks, skip connections are established.Three-dimensional UTE cones sampling was performed using an echo time (TE) of 32 μs, repetition time (TR) of 20 ms, and flip angles (FAs) of 5°, 10°, 20°, and 30°, respectively, for UTE-T1 mapping. For UTE-T1rho mapping, the scan parameters were TR=500ms, flip angle=10°, and 7 spin-lock times (TSLs)=0, 12, 24, 36, 48, 72, and 96 ms. Subsequently, image registration was performed to minimize inter-scan motion. M0, UTE-T1, UTE-T1rho, and B1 maps were derived via non-linear fitting using the Levenberg‐ Marquardt algorithm. ROIs for the cartilage and meniscus area were labeled with homemade matlab code by three experienced radiologists4,5.
A total of 1056 slice images from 44 subjects (including healthy volunteers and patients with different degrees of OA) were used for model training, and 144 images of six additional subjects were used for model validation. The DCNNs were implemented with pytorch 1.1.0 on a workstation with a Nvidia GTX 1080 Ti (11 GB GPU memory).
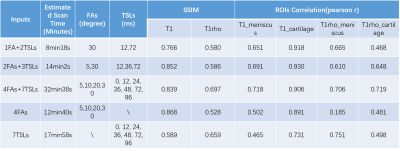
Several experiments of the combination of the input images were performed, including full data with all four FAs and 7 TSLs (4 FAs+7 TSLs), and partial data with 2FAs + 3TSLs, and 1FA + 2TSLs. All FAs and TSLs were carefully selected to achieve the best performance. As the input image number changed for each experiment, the MSMQ-Net input channel was modified accordingly and trained separately with 300 epochs. Detailed information can be found in Table 1.
Structural Similarity (SSIM) score is applied to compare the generated UTE-T1 and UTE-T1rho images with the images generated from full data (4FAs+7TSLs) with the traditional fitting method6. Pearson correlation was performed to evaluate the relationship between the predicted values from the MSMQ-net and the ground truth for all regions of interest (ROIs).
Results and Discussion
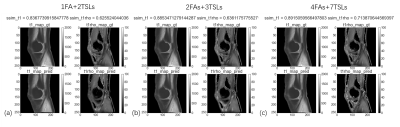
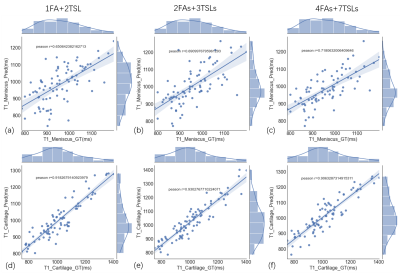
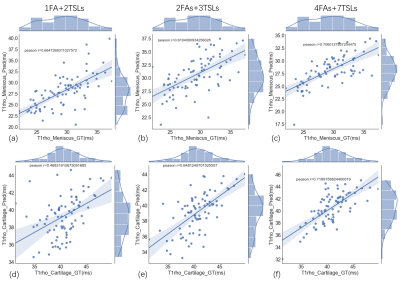
A typical example of predicted qMRI mapping from different combination could be found in Figure 2. Compared with full data (4FAs+7TSLs) as the input, both 1FA+2TSLs and 2FAs+3TSLs could obtain similar predicted UTE-T1 mapping of the knee, but a noticeable decrease of the ssim score in UTE-T1rho mapping.Figure 3 shows the correlation studies between the predicted UTE-T1 maps of the meniscus and cartilage regions, which indicate that the correlation r is a bit decreased for the meniscus due to the reduction of the input numbers, while the correlation r of the cartilage ROIs is not. Figure 4 shows the correlation studies between the predicted UTE-T1rho maps of the meniscus and cartilage regions, where the r of the cartilage significantly decreases due to the input signals’ reduction.
All the experimental results are summarized in table 1. It could be concluded that the scan time of 1FA+2TSLs is 8min18s, which is about 25% of the full input of 4FAs + 7TSLs, but the ssim score of UTE-T1 and UTE-T1rho is kept as 90% of its full input, which implies the possibility of further accelerating the scan with MSMQ-Net with fewer inputs. It is also quite interesting to notice that for the 4FAs, the predicted ssim score of UTE-T1 mapping is the highest, while the ssim score of UTE-T1rho mapping is quite low. For the 7TSLs input, the situation is vice versa.
Conclusion
In this study, different combinations of the UTE MRI images were tested as the input of the MSMQ-Net. The results demonstrated that with 25% of full inputs, the performance remains at a reasonable level (90%), indicating the possibility of accelerating the scan with MSMQ-Net.Acknowledgements
The authors acknowledge grant support from the National Institutes of Health (R01AR062581, R01AR068987, R01AR075825, R01AR079484, and RF1AG075717 and R21AR075851), VA Clinical Science and Rehabilitation Research and Development Services (Merit Awards I01CX001388, I01CX002211, and I01RX002604), and GE Healthcare.References
1. Alexander Selvikvåg Lundervold, Arvid Lundervold.An overview of deep learning in medical imaging focusing on MRI,Zeitschrift für Medizinische Physik,Volume 29, Issue 2, 2019,Pages 102-127.
2. Lu X, Ma YJ, Saeed J, Jang H, Xue YP, Zhang XD, Wu M, Gentili A, Hsu CN, Chang EY, Du J. Deep CNNs with Physical Constraints for simultaneous Multi-tissue Segmentation and Quantification (MSQ-Net) of Knee from UTE MRIs. ISMRM 2021.
3. Lu X, Ma YJ, et al. Deep CNNs with Physical Constraints for simultaneous Multi-tissue Segmentation and Multi-parameter Quantification (MSMQ-Net) of Knee. International Society for Magnetic Resonance in Medicine(ISMRM) 2022, 07-15 May, London, England, United Kingdom
4. Ma YJ, Lu X, Carl M, Zhu Y, Szeverenyi NM, Bydder GM, Chang EY, Du J. Accurate T1 mapping of short T2 tissues using a three-dimensional ultrashort echo time cones actual flip angle imaging-variable repetition time (3D UTE-Cones AFI-VTR) method. Magn Reson Med. 2018 Aug;80(2):598-608.
5. Ma YJ, Carl M, Searleman A, Lu X, Chang EY, Du J.3D adiabatic T1ρ prepared ultrashort echo time cones sequence for whole knee imaging. Magnetic resonance in medicine 80 (4), 1429-1439. 2018
6. Wang Z, Bovik A.C., Sheikh H. R. and Simoncelli E. P.Image quality assessment: From error visibility to structural similarity, IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600-612, Apr. 2004.
Figures