4517
Improved Robustness for Deep Learning-based Segmentation of Perfusion CMR Using Data Adaptive Uncertainty-guided Spatiotemporal Analysis
Dilek Mirgun Yalcinkaya1,2, Khalid Youssef3, Bobak Heydari4, Subha Raman3,5, Rohan Dharmakumar3,5, and Behzad Sharif1,3,5
1Laboratory for Translational Imaging of Microcirculation, Indiana University (IU) School of Medicine, Indianapolis, IN, United States, 2Elmore Family School of Electrical and Computer Engineering, Purdue University, West Lafayette, IN, United States, 3Krannert Cardiovascular Research Center, IU School of Medicine/IU Health Cardiovascular Institute, Indianapolis, IN, United States, 4Stephenson Cardiac Imaging Centre, Department of Cardiac Sciences, University of Calgary, Calgary, AB, Canada, 5Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States
1Laboratory for Translational Imaging of Microcirculation, Indiana University (IU) School of Medicine, Indianapolis, IN, United States, 2Elmore Family School of Electrical and Computer Engineering, Purdue University, West Lafayette, IN, United States, 3Krannert Cardiovascular Research Center, IU School of Medicine/IU Health Cardiovascular Institute, Indianapolis, IN, United States, 4Stephenson Cardiac Imaging Centre, Department of Cardiac Sciences, University of Calgary, Calgary, AB, Canada, 5Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation
We proposed and validated Data Adaptive Uncertainty-Guided Spatiotemporal (DAUGS) analysis that leverages the data-driven uncertainty map of the segmentation contours among a pool of trained deep neural networks (DNNs) and automatically selects the segmentation result with the highest level of certainty. Our results suggest that proposed DAUGS and standard DNN-based analysis demonstrated on-par performance on the internal test set which is from the same institution as training set and acquired with FLASH sequence. In contrast, DAUGS analysis considerably outperformed DNN-based analysis on the external test set which was acquired with a bSSFP pulse sequence at a different institution, demonstrating the improved robustness of the proposed method despite limited training data.Introduction
Fully automatic analysis of first-pass perfusion (FPP) myocardial MR datasets enables rapid and objective reporting of stress/rest studies in patients with suspected ischemic heart disease [1-3]. With deep learning-based approaches, training well-generalizable deep neural network (DNN) models that, despite having a limited training dataset, are robust across different sites and data-acquisition protocols is an ongoing challenge. Previous work has shown that a “sliding patch” approach for analysis of FPP images generates a data-driven pixelwise “uncertainty map” as a byproduct of the segmentation process [4,5].Methods
We propose to leverage the data-driven uncertainty map (U-map) among a pool of multiple trained DNNs (all with the same 3D U-Net architecture but trained with different parameter initializations) to perform Data Adaptive Uncertainty-Guided Spatiotemporal (DAUGS) analysis as in Fig 1, and automatically select the “best” segmentation result with the highest level of certainty in a data-adaptive manner by computing total per-pixel energy Upp of the U-maps (Step 4 in Fig 1). FPP data from 106 patients with suspected ischemia and 14 healthy subjects acquired at 3T from two sites were used: (1) an internal dataset acquired using a saturation recovery (SR) prepared FLASH sequence, and (2) an external dataset acquired with SR-prepared SSFP (Fig 2). Training of DNNs used the data from a subset of the internal dataset (330 stress/rest FPP image series; 90% females). Performance of the proposed DAUGS vs. standard DNN-based analysis was evaluated on a small subset of the internal dataset which has no overlap with the training data and the entire external dataset (120 stress FPP image series; 25% females).Results
Fig 3 summarizes the Dice-score comparison of the proposed DAUGS analysis approach vs. standard DNN-based analysis. The “standard” DNN-based analysis approach refers to the conventional DNN training approach in which a single model is selected during the validation process. For the internal dataset, our proposed method and standard approach showed a comparable performance (p >0.5). However, on the external dataset, ours significantly outperformed the standard approach (Dice: 0.885 ± 0.040 vs. 0.849 ± 0.065, p < 0.01). Fig 4 shows a challenging case from the external test set with diffuse stress-induced ischemia in all 3 short-axis slices and LV hypertrophy which renders the mid slice (acquired end systole) very challenging to segment. The segmentation chosen by the proposed approach performs well with a mean Dice score of >0.90, whereas the standard approach fails to segment to mid slice. Overall, the number of “failed” segmentations (discontiguous contours) was markedly lower for the proposed method (< 1% vs. 5%).Conclusion
Our proposed data-adaptive approach for analysis of FPP datasets offers the flexibility to choose the final segmentation result from a pool of candidate solutions based on the uncertainty level detected by the trained spatiotemporal DNNs. Our results demonstrate that the proposed DAUGS analysis approach improves the generalization ability of DNN-based analysis despite the limited training data, which in turn has the potential to enable automatic analysis of perfusion CMR datasets with improved robustness to variations in the data acquisition protocol (SR- FLASH vs. SR-SSFP), sequence parameters, or site location.Acknowledgements
No acknowledgement found.References
1. Xue H, Davies RH, Brown LA, Knott KD, Kotecha T, Fontana M, et al. Automated inline analysis of myocardial perfusion MRI with deep learning. Radiology: AI 2020;2(6):e200009.2. Scannell CM, Veta M, Villa ADM, Sammut EC, Lee J, Breeuwer M, et al. Deep-Learning-Based Preprocessing for Quantitative Myocardial Perfusion MRI. JMRI 2020;51(6):1689-96.
3. Hann E, Popescu IA, Zhang Q, Gonzales RA, Barutçu A, Neubauer S, et al. Deep neural network ensemble for on-the-fly quality control- driven segmentation of cardiac MRI T1 mapping. Medical image analysis. 2021 Jul 1;71:102029.
4. Yalcinkaya DM, Youssef K, Heydari B, Zamudio L, Dharmakumar R, Sharif B. Deep Learning-Based Segmentation and Uncertainty Assessment for Automated Analysis of Myocardial Perfusion MRI Datasets Using Patch-Level Training and Advanced Data Augmentation. Proceedings of IEEE Engineering in Medicine & Biology Society (EMBC); 2021;4072-78.
5. Lu MY, Williamson DF, Chen TY, Chen RJ, Barbieri M, Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nature Biomedical Engineering 2021;5(6):555-70.
Figures
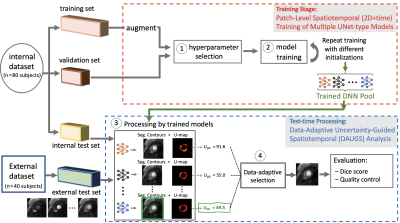
Figure 1. Description of the proposed approach and two datasets used for training and testing. Having split the internal dataset into three subsets, hyperparameter selection was done. Steps 1 & 2: All models were trained using a patch-level approach [4,5] and a pool of trained deep neural networks (DNNs) was obtained by varying the parameter initializations. Step 3: At test time, each DNN in the pool provides a segmentation and a pixel-wise uncertainty map (U-map). Step 4: Our data-adaptive approach selects the segmentation result with the lowest uncertainty (Upp).
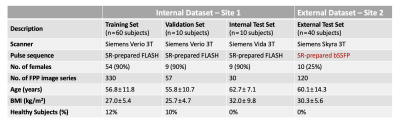
Figure 2. Table summarizing the characteristics of the internal and external datasets (2 sites) used in this study. There were notable differences between the internal dataset and external dataset in terms of subject characteristics, data acquisition protocol (choice of SR- prepared FLASH vs. SR-prepared bSSFP) and pulse sequence parameters (SR time, resolution, flip angle, etc.). Training of the DNNs used a subset of the internal dataset and there was no overlap between the training data and testing data.
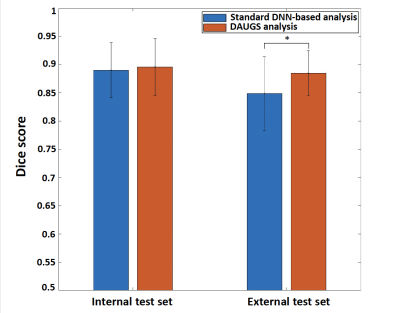
Figure 3. Comparison of the segmentation performance of the proposed DAUGS analysis approach vs. standard DNN-based analysis. For the internal test set, the two approaches performed similarly (p > 0.5) whereas, on the external dataset, the proposed method significantly outperformed the standard approach (Dice: 0.885 ± 0.040 vs. 0.849 ± 0.065, p < 0.01). This shows improved generalization ability for the proposed DAUGS analysis approach vs. standard DNN-based analysis despite the limited training data.
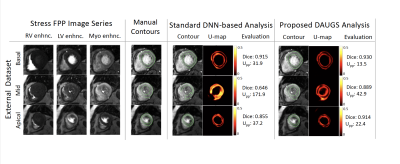
Figure 4. Segmentation results for a challenging case from the external test set. In the mid slice, in particular, the small LV and RV bloodpools makes the detection of subendocardial and septal epicardial contours challenging even for an expert reader. The segmentation chosen by DAUGS analysis performs with a mean Dice score of >0.90 across 3 slices, whereas the standard DNN-based analysis performs notably worse for the apical slice and fails segment the mid slice (also reflected in the corresponding U-maps and Upp values).
DOI: https://doi.org/10.58530/2023/4517