4515
Self-Supervised Learning for Perivascular Spaces segmentation with enhanced contrast knowledge1University of Southern California, Los Angeles, CA, United States, 2NeuroScope Inc., Scarsdale, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation, self-supervised learning
Due to the absence of isotropic T2w modality in clinical datasets, it is challenging to enhance the PVS contrast using multiple neuroimage modalities. To overcome this issue, in this work we introduced using self-supervised pre-trained model in the enhanced PVS contrast image space to improve the downstream model segmentation performance when solely using T1w as the training data. The experiment results showed that the proposed method increased segmentation accuracy compared to the model trained from scratch using T1w modality and resulted in faster training and less required training data volume.Introduction
There is an increasing interest in the role of PVS morphology alteration in cerebrospinal fluid circulation and clearance of cerebral waste products, which has inspired automatic PVS segmentation method developments [1]. Sepehrband F. et al. [2] proposed an algorithmic image processing method and PVS enhanced contrast modality (EPC) to achieve reliable PVS segmentation results. However, the absence of isotropic T2w modality in many clinical datasets limit the benefit of using EPC modality. Self-supervised learning [3] has been introduced into the medical imaging analysis field in recent years to tackle the heavy dependency on the largely manually labeled datasets for the training purpose of the deep learning models [4]. Based on the successful implementations of self-supervised learning model in the segmentation tasks, we tested this training paradigm in the PVS segmentation task. Because of the transfer learning nature of self-supervised learning, we propose the method to pre-train the model in self-supervision on PVS EPC space and transfer the knowledge to the downstream PVS segmentation task where only T1w modality was available.Methods
The main purpose of the pre-text task was teaching the model to learn the intrinsic knowledge of the input modality. In our experiment, pretext task was designed to teach the model to segment bright tubular voxels (PVS in this case) in the EPC modality with automaticly generated training target using Frangi filter algorithm [2]. For the downstream task, we only used T1w modality and quality controlled PVS segmentation maps as the training targets. This training paradigm aimed to teach the model to learn PVS distribution in the high PVS contrast modality by the self-supervised learning process, then train the model on the lower PVS contrast modality with the human curated PVS maps. For the pre-text task training, we used 2000 subjects from the HCP dataset [5] with EPC modality and Frangi filter generated PVS map. For the downstream task training, we used 194 subjects with T1w modality and quality controlled (QC) PVS map. To verify the benefits of self-supervised learning for the PVS segmentation task, we used 6, 27, 87, 174 subjects correspondingly for the downstream task training which included subjects from the three cohorts to balance the data. 20 subjects were used as the testing dataset for each experiment. We utilized SC-GAN [6] as the backbone model and followed its implementation. We also implemented binary focal-loss [7] as the loss function as follows: where q is the target PVS binary map and p is the output probability map of SC-GAN, assigns high weights to voxels whose prediction errors was larger than 0.4, so the model could focus more on the voxel predictions with large errors and less on the voxel predictions with small error. The final threshold to binarize the output probability map of SC-GAN is 0.5. Patches with size of 128*128*128 were extracted from each subject as the training data. Data augmentation was conducted by rotating the original image 90 degree in two directions along one of the x, y and z axes in the 3D image coordinates.Results and discussion
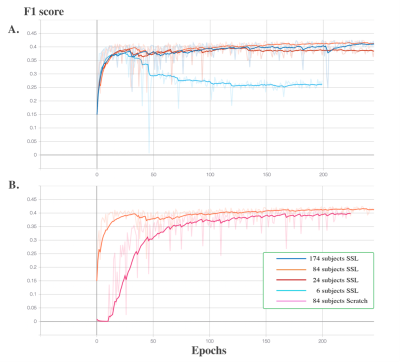
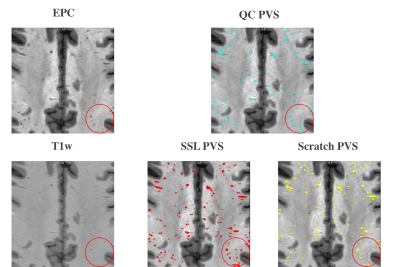
F1 score were used as the evaluation metric for each downstream experiment. In Figure 1. A., the learning curves of the four experiments using 6, 27, 87 and 174 subjects reach the plateaus of 0.375, 0.39, 0.42 and 0.42 F1 score, respectively. The experiment on using 6 subjects as the training set resulted in severe overfitting. An additional experiment using 87 subjects as the training set was conducted and the model was trained from scratch. In Figure 1. B., the learning curve of the model trained from scratch reached the plateau of 0.40, while the self-supervised trained model reached the plateau of 0.42. In addition, Figure 1. A. shows that the small size (84 subjects) of the training dataset could still reach the same segmentation accuracy compared to the large size (174 subjects) dataset for the downstream task, which shows the advantage of transfer learning. Figure 1. B. shows the improved segmentation accuracy of self-supervised trained model compared to the model trained from scratch with the same amount of humanly curated training subjects, which is the advantage of incorporating a pre-text task trained with high PVS contrast EPC modality. In Figure 2, the region in red circle represents the PVS which have less contrast in T1w compared to EPC. The model trained using proposed self-supervised learning method could capture the PVS with weak contrast in T1w (second image in the second row), while the model trained from scratch missed couple of weak contrast PVS (third image in the second row).Conclusion
Here we proposed a self-supervised learning method to tackle the PVS segmentation task and to test the possibility of overcoming the limitation caused by the absence of isotropic T2w modality in the clinical datasets. Our pre-text model was trained on 2000 HCP subjects and the experiment results showed the advantage of using pre-text trained model to improve the segmentation performance on downstream task solely using T1w as the input data. Our future work will focus on verifying the effectiveness of our HCP data trained pre-text model on the downstream tasks with other clinical neuroimage datasets.Acknowledgements
The research reported in this publication was supported by the National Institute of Mental Health of the NIH under Award Number RF1MH123223, R01AG070825, R01NS128486, and R41AG073024.References
[1] G. Barisano et al., “Imaging perivascular space structure and function using brain MRI,” Neuroimage, vol. 257, no. May, p. 119329, 2022, doi: 10.1016/j.neuroimage.2022.119329.
[2] F. Sepehrband et al., “Image processing approaches to enhance perivascular space visibility and quantification using MRI,” Sci. Rep., vol. 9, no. 1, pp. 1–12, 2019, doi: 10.1038/s41598-019-48910-x.
[3] C. Doersch and A. Zisserman, “Multi-task Self-Supervised Visual Learning,” in IEEE international conference on computer vision, 2017, pp. 2051–2060.
[4] W. Bai et al., “self-supervised learning for cardiac MR Image Segmentation by Anatomical Position Prediction,” in Proceeding of the International Conference on Medical Image Computing and Computer Assisted Interventions, 2019, pp. 541–549, doi: 10.1007/978-3-030-32245-8.
[5] D. C. Van Essen et al., “The Human Connectome Project: A data acquisition perspective,” Neuroimage, vol. 62, no. 4, pp. 2222–2231, 2012, doi: 10.1016/j.neuroimage.2012.02.018.
[6] H. Lan, A. W. Toga, and F. Sepehrband, “Three-dimensional self-attention conditional GAN with spectral normalization for multimodal neuroimaging synthesis,” Magn. Reson. Med., vol. 86, no. 3, pp. 1718–1733, 2021, doi: 10.1002/mrm.28819.
[7] T.-Y. Lin, P. Goyal, R. Girshick, K. He, and P. Dollár., “Focal Loss for Dense object Detection,” in In Proceedings of the IEEE international conference on computer vision, 2017, pp. 2980–2988, doi: 10.1109/ICAICTA49861.2020.9428882.
Figures