4514
Semi-supervised segmentation for 3D medical image based on contrast learning1Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Data Processing, MRI medical segmentation
Semi-supervised segmentation, using large amounts of unlabeled data and small amounts of labeled data, has achieved great success. This paper proposes a semi-supervised segmentation method based on consistent learning and contrast learning. It mainly uses a mean-teacher framework to add consistency losses and contrast losses based on multiscale features to minimize the distance of model responses under different disturbance inputs. In addition, mean square error loss was used to alternately minimize the gap between the teacher and student models. In 3D left atrium data, a Dice coeffivient of 0.8970 was obtained, which was superior to other methods.Introduction
The structure of the left atrium is essential information for clinicians to diagnose and treat atrial fibrillation[1]. Medical image segmentation methods have achieved great success, but they rely on a large amount of annotated data, which is difficult to obtain for medical images. Semi-supervised segmentation solves this problem by using large amounts of unlabeled data and small amounts of labeled data[2]. The most successful semi-supervised learning approaches are based on consistency learning that minimizes the distance between model responses obtained from perturbed views of the unlabeled data[3]. Moreover, contrast learning has been proven to be an effective method of unsupervised learning[4]. This paper proposes a semi-supervised learning method based on the mean-teacher framework, which combines consistent learning and contrast learning, and uses the mean-square error loss to alternately optimize the model results. The results show that the proposed method is superior to other methods.Materials and methods
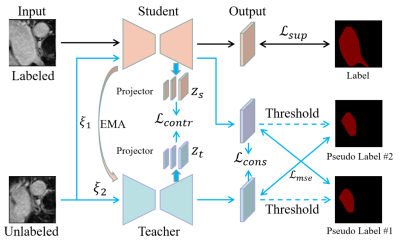
Dataset: We demonstrated our approach on an MRI dataset, namely, left atrium(LA)[5]. The left atrium dataset contains a total of 100 3D MRI volumes. Following previous papers[5,6], we used the same data division, 80 for training and 20 for testing.Network: We proposed a mean-teacher[7] semi-supervised learning method based on contrast learning, the overall structure is shown in Fig. 1, with the same structure of the student and teacher models, both are V-Net[8]. The input data of the network contains both labeled and unlabeled data, with labeled data used only for the student model, and is optimized using cross-entropy and dice, denoted as $$${Loss}_{sup}$$$, and we pass the parameters of the student model to the teacher model by the exponential moving average (EMA)[9] method in different training steps. The unlabeled data were used for the teacher model and the student model, and different Gaussian noise, denoted $$$\xi_1$$$ and $$$\xi_2$$$, were added to the two model inputs. The outputs of both were first obtained as consistency loss $$${Loss}_{cons}$$$, and then after thresholding, both were alternately obtained as the mean-square error loss $$${Loss}_{mse}$$$. Moreover, we add contrast loss between the teacher-student model by outputting the feature maps of each stage of the decoding process of the teacher-student model as multiscale features after the projector, and then calculating the contrast loss $$${Loss}_{contr}$$$. Finally, we obtain the unsupervised loss $$${Loss}_{unsup}={Loss}_{contr}+{Loss}_{cons}+{Loss}_{mse}$$$.
Loss function: We use cross entropy and the Dice loss function to optimize the labeled data. Denoted as: $$${Loss}_{Dice}(y, \hat{y})=1-\frac{2 \sum_i^N y_i \hat{y}_i}{\sum_i^N y_i+\sum_i^N \hat{y}_i+\varepsilon}$$$ and $$${Loss}_{CE}(y, \hat{y})=-\sum_{i=1}^N y_i \log \hat{y}_i+\left(1-y_i\right) \log \left(1-\hat{y}_i\right)$$$, where $$$y_i$$$ is the label and $$$\hat{y_i}$$$ is the predicted probability of the i-th voxel. N is total number of voxels, and $$$\varepsilon$$$ is a small constant to avoid the zero division. $$$\operatorname{Loss}_{Sup }=\operatorname{Loss}_{Dice}(y, \hat{y})+\operatorname{Loss}_{\mathrm{Ce}}(y, \hat{y})$$$. For the unsupervised data, we used the mean-square error loss, which is defined as: $$$\operatorname{Loss}_{mse}(y, \hat{y})=\frac{\sum_{\mathrm{i}=1}^{\mathrm{N}}\left(y_i-\hat{y}_i\right)^2}{\mathrm{~N}}$$$, and the consistency loss $$${Loss}_{cons}$$$ for unlabeled data is calculated by the mean-squared error: $$${Loss}_{cons}=\mathcal{F}\left(P_u^S, P_u^T\right)$. $P_u^S$$$ and $$$P_u^T$$$ denote the predicted values of the student model and the teacher model for the unlabeled data, respectively, and $$$\mathcal{F}$$$ is the mean-squared error. After adding different noises to the unlabeled data, the projection features output by the student model and the teacher model are $$$z_i$$$ and $$$z_j$$$, respectively. Then, the loss for a positive pair of examples (i, j) is defined as $$${Loss}_{\text {contr }(i, j)}=-\log \frac{\exp \left(\operatorname{sim}\left(z_i, z_j\right) / \tau\right.}{\sum_{k=1}^{2 N} \mathbb{L}_{\{k \neq i\}} \exp \left(\operatorname{sim}\left(z_i, z_k\right) / \tau\right)}$$$, where N is the number of examples, and $$$\mathbb{L}_{\{k \neq i\}} \in\{0,1\}$$$ is an indicator function evaluating to 1 iff $$$k \neq i$$$ and $$$\tau$$$ denotes a temperature parameter, $$$\operatorname{sim}(u, v)=\frac{u^{\mathrm{T}} \mathrm{v}}{\|u\| \cdot\|v\|}$$$. The final loss is computed across all positive pairs, both (i, j) and (j, i), $$${Loss}_{\text {contr }}={Loss}_{{contr }(i, j)}+{Loss}_{{contr}(i, j)}$$$. $$${ Loss }_{unsup}={Loss}_{contr}+{Loss}_{cons}+{Loss}_{ mse}$$$, and the final total loss is $$$Loss={Loss}_{sup}+{Loss}_{unsup}$$$.
Results
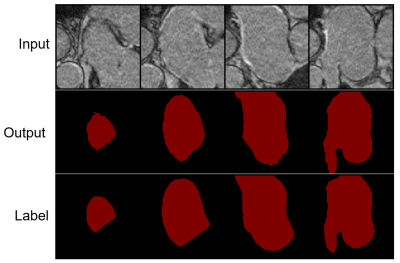
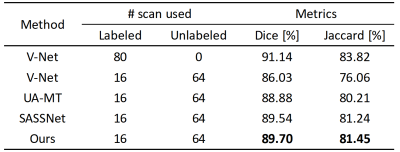
We validated the proposed method on 3D left atrial data. Table 1 shows the quantitative segmentation results of the proposed network in this paper. On both the Dice and Jaccard metrics, our method achieved the best segmentation results. Fig. 2 shows the segmentation visualization results for the four cases.Conclusion
In this paper, we proposed a semi-supervised segmentation method based on contrast learning, which achieves good segmentation results on the 3D left atrial dataset and outperforms other contrast algorithms.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81871441), and the Shenzhen Excellent Technological Innovation Talent Training Project of China (RCJC20200714114436080).References
1. Guglielmo M, Baggiano A, Muscogiuri G, et al. Multimodality imaging of left atrium in patients with atrial fibrillation[J]. Journal of Cardiovascular Computed Tomography, 2019, 13(6): 340-346.
2. Ouali Y, Hudelot C, Tami M. Semi-supervised semantic segmentation with cross-consistency training[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020: 12674-12684.
3. Chen X, Yuan Y, Zeng G, et al. Semi-supervised semantic segmentation with cross pseudo supervision[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021: 2613-2622.
4. Chen T, Kornblith S, Norouzi M, et al. A simple framework for contrastive learning of visual representations[C]//International conference on machine learning. PMLR, 2020: 1597-1607.
5. Yu L, Wang S, Li X, et al. Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 605-613.
6. Li S, Zhang C, He X. Shape-aware semi-supervised 3D semantic segmentation for medical images[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020: 552-561.
7. Tarvainen A, Valpola H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results[J]. Advances in neural information processing systems, 2017, 30.
8. Milletari F, Navab N, Ahmadi S A. V-net: Fully convolutional neural networks for volumetric medical image segmentation[C]//2016 fourth international conference on 3D vision (3DV). IEEE, 2016: 565-571.
9. Tarvainen A, Valpola H. Weight-averaged consistency targets improve semi-supervised deep learning results. CoRR abs/1703.01780[J]. arXiv preprint arXiv:1703.01780, 2017, 1(5).