4512
Efficient Fetal Brain Segmentation according to the Point Spread Function of MRI1Zhejiang University, Hangzhou, China, 2Biomedical Engineering, University of Virginia, Virginia, VA, United States, 3Department of Radiation Oncology, City of Hope National Center, Los Angeles, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Fetus, Fetal Brain MRI,Point Spread Function
High apparent resolution of fetal MRIs is provided by slice-to-volume reconstruction pipelines widely. However, the physical resolution of the fetal brain is lower than that. Therefore, we hypothesize that fetal brain segmentation can be performed based on downsampled fetal brain MRI according to its point spread function. In this work, 150 adult brain and 80 fetal brain MRIs were used to validate hypothesize. Using downsampled fetal data with factor of 4, a highly efficient segmentation model achieved similar segmentation accuracies compared to original data, which demonstrated that segmentation models can be developed based on PSF.
INTRODUCTION
Fetal brain MRI provides essential information about early brain developments 1. Fetal MR images are acquired in 2D slices and reconstructed to 3D volumes to reduce the artifacts from fetal and maternal movements. To improve the resolution in the slice direction and to reduce the motion through imaging slices, the stack-of-slices can be acquired in three orthogonal directions and 3D images can be reconstructed using slice-to-volume algorithms 2–4. However, there has been a consensus in the MRI field that the PSF of 3D reconstruction images can not be higher than the in-plane PSF of acquired 2D images 5. Therefore, the resulting fetal brain MRI has a high apparent resolution but a low physical resolution, which results in unnecessary computational cost in segment. In this work, we proposed that similar segmentation accuracies can be achieved by downsampling the apparent resolution of fetal MRI to its PSF.METHODS
Dataset: The first dataset was obtained from the ADNI database (adni.loni.usc.edu, RRID:SCR_003007). 150 subjects of the ADNI3 study 6 were selected, which was segmented into four categories using SPM12. The second dataset was the fetal brain MRI from the FeTA Challenge of MICCAI 2021 7, which had 80 fetal brain T2-weighted MR images with an apparent resolution of 0.5mm isotropic but PSF is 2mm.SSIM 8 was used to calculate the gap between the original and the undersampled image, which quantify the information in high PSF and high apparent resolution images.
Performances of segmentation models were evaluated on downsampled images. For the ADNI3 dataset, the downsampling step reduced the PSF. But for fetal brain MRI, the PSF would remain similar, since the original PSF was lower than its apparent resolution. Three studies were performed in this work.
(1) Effects of the PSF: ADNI3 images and labels were downsampled with factors of 4/3, 2, and 4, which resulted in images with PSFs of 4/3mm, 2mm, and 4mm, respectively. The nnUnet 9 and a modified 3D UNet 10 (mUNet) were trained using the original and undersampled data.
(2) Effects of the apparent resolution: Based on (1), the downsampled data (factor of 4) were upsampled to the original matrix size using linear interpolation, which is high apparent resolution data. The mUNet was trained on these data.
(3) Downsampled fetal MRI according to its PSF: The fetal images and labels were downsampled by factors of 4/3, 2, and 4 in each dimension like Study 1, while its PSF remains the same (≈2mm). The accuracies were evaluated with manual label, which need upsampling predict segmentation.
The mUNet was built based on the backbone of 3D Unet. Patch-based train and inference were performed. The loss function was binary cross-entropy, the performance was evaluated by dice coefficient.
RESULTS
The SNR of the ADNI3 and fetal MRI were 15.2±5.7 and 18.8±3.5. When MR images were downsampled by a factor of 4, the SSIM to the original images was reduced to 0.69 in the ADNI data, while that was reduced to 0.9 slightly in the fetal MRI.Figure 1 shows the image's structural information loss after sampling. Compared to ADNI images, fetal images did not lose additional structures during downsampling, which indicated a low physical resolution in its original images.
Undersampled ADNI images resulted in significantly reduced segmentation accuracy in both nnUNet and mUNet, Table 1. When high apparent resolution MRIs were provided using linear interpolation, similar segmentation accuracy was found compared to the undersampled MRI, without significant differences, Table 1.
More importantly, when the fetal brain MRI was downsampled by factors up to 4, segmentation accuracies of nnUNet and mUNet were slightly compromised by 3.1% and 1.7% with the undersampled data, Table 2. It indicated the original low physical resolution in fetal MRIs and confirmed the feasibility of the proposed method. In Figure 2, accurate segmentations were achieved with downsampled fetal images both at early and late gestational ages. Figure 3 shows the performance drops in segmentation accuracy, when ADNI and Fetal data were undersampled.
Discussion and Conclusion
This work demonstrated that the segmentation models can be developed accurately and efficiently based on the PSF of fetal brain MRI. By downsampling the fetal brain MRI to its PSF, the computational cost can be reduced largely, while the segmentation accuracy can be preserved. The proposed work is an independent data preprocesing module. Therefore, it is compatible to improve the performance of segmentation methods in general.There are a few possible reasons for the slightly compromised performance, including more accurate estimation of the PSF and possible subjective bias in manual labeling between raters 11-12.
Acknowledgements
This work is supported in part by the Alzheimer's Association through AARF-18-566347, the MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University, Zhejiang Provincial Natural Science Foundation of China under Grant No. LGJ22H180004, 2020R01003, and 2022C03057, and Alibaba Cloud.
References
Figures
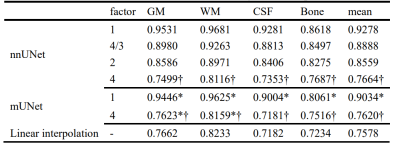
Table 1 Dice coefficients of ConvNets using downsampled ADNI3. * stands for significant differences (p<0.0001) in the dice coefficients between the nnUNet and the mUNet models at a downsampling factor of 1 or 4 in each dimension. † stands for significant dice differences (p<1e-14) between the original data and the downsampled data by a factor of 4 in each model.
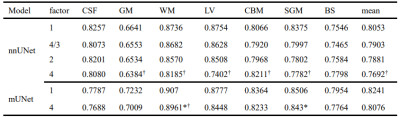

Figure 1. PSF and apparent resolution on selected adult brain MRI (top) and fetal brain MRI at early (middle) and late (bottom) gestational ages. (a) high PSF image. (b) downsampled image with a factor of 4 and then refill to the original matrix size using a pixel padding method. The high apparent resolution image is shown in (c), in which the undersampled image was refill to the original matrix size using the linear interpolation for adult brains and they are the original images provided by the reconstruction in fetal brain MRIs.
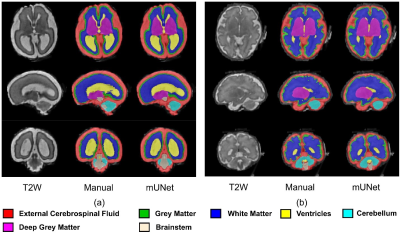
Figure 2. Selected segmentations of fetal brain MRIs at early (a) and late (b) gestational ages on undersampled data by a factor of 4.
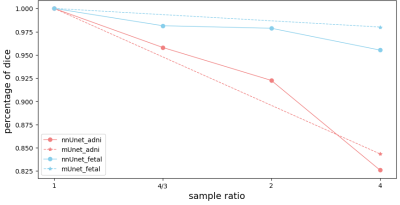
Figure 3. Dice coefficients of ConvNets using downsampled. Compared to the unsampled method, the decreased ratio of dice coefficients for the downsampled method.