4511
The Yale Glioma Dataset: Developing An Open Access, Annotated MRI Database1Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States, 2University of Düsseldorf, Düsseldorf, Germany, 3University of Ulm, Ulm, Germany, 4Leipzig University, Leipzig, Germany, 5Visage Imaging, Düsseldorf, Germany, 6Pathology, Yale School of Medicine, New Haven, CT, United States, 7Therapeutic Radiology, Yale School of Medicine, New Haven, CT, United States, 8Neurology, Yale School of Medicine, New Haven, CT, United States
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence, Glioma
Recent development of Machine Learning (ML) tools for analysis of CNS tumors demonstrates great potential benefit to research and clinical practice but has been hindered by a lack of external validation. There is a critical need for open access to large individual hospital-based datasets with expert annotations. Here, we present the Yale Glioma Dataset, a database of 1,033 patients featuring annotated segmentations on FLAIR and T1 post-gadolinium, tumor grading and classification, and further clinical information. Open access of this database will support the development and validation of new AI algorithms for glioma detection and segmentation.Introduction
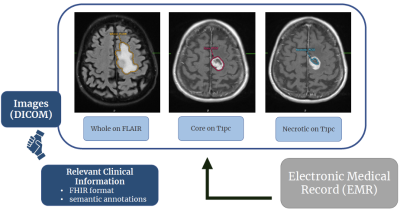
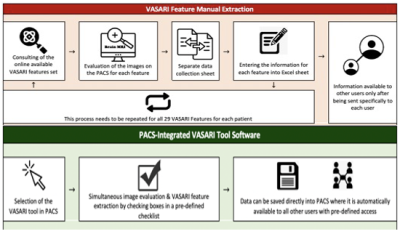
Gliomas are the most common primary brain malignancy, comprising about 80% of malignant primary CNS tumors in the US4. Machine learning (ML) has demonstratedtremendous progress in predicting glioma grade (classified by the World Health Organization into grades 1-4)5 and molecular subtypes based on radiomic analysis of MR images. However, translation of AI algorithms into clinical practice is significantly limited by lack of large individual hospital-based datasets with expert annotations. Current methods for generation of annotated imaging data are significantly limited due to inefficient imaging data transfer, complicated annotation software, and time required for experts to generate ground truth information. We incorporated AI tools for auto-segmentation of gliomas into PACS that is used at our institution for reading clinical studies. We developed a clinical workflow for annotation of images and generation of volumetric segmentations in neuroradiology. Here, we present the Yale Glioma Dataset, an open access database that includes annotated images of 1,033 gliomas with associated clinical outcome information.Methods
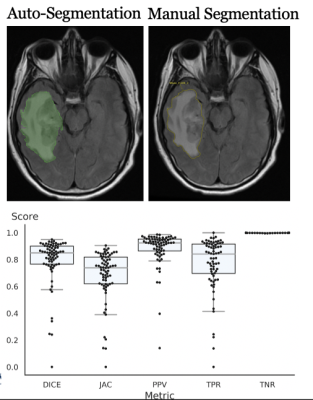
Our database includes 1,033 patients (including 595 Grade 4 gliomas) with FLAIR images, T1 post-gadolinium images, molecular subtype classification, tumor classification, and corresponding survival data. Patients were identified within the Yale Radiation Oncology Registry (2012-2019) after IRB approval. Volumetric segmentations of whole tumor, enhancing tumor, and peritumoral edema were generated in a semiautomatic workflow using a UNETR algorithm trained on BRaTS 2021 dataset and internally validated on a Yale adult/pediatric glioma dataset. Segmentations were validated by a board-certified neuroradiologist, after which PACS-embedded PyRadiomics was used for direct feature extraction.Results
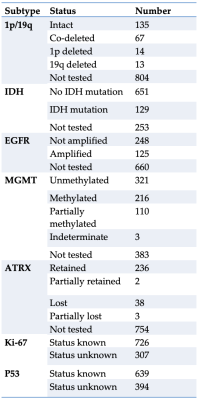
In 7 Months (05/2021 - 08/2021, 03/2022 - 06/2022) segmentations and annotations were performed in 1,033 patients (429 female, 604 male, mean age 52.6 years). Dataset includes 595 Grade 4 Gliomas (96 Grade 3, 105 Grade 2, 45 Grade 1, 192 unknown). Molecular subtypes include IDH (129 mutated, 651 wildtype, 253 unknown), 1p/19q (94 deleted or co-deleted, 135 intact, 804 unknown), MGMT promotor (216 methylated, 110 partially methylated, 321 unmethylated, 386 unknown), EGFR (125 amplified, 248 not amplified, 660 unknown), ATRX (43 mutated, 236 retained, 754 unknown), Ki-67 (726 known, 307 unknown) and p53 (639 known, 394 unknown).Discussion
The Yale Glioma Dataset includes a large cohort of glioma patients with annotated FLAIR and T1 post-gadolinium images as well as associated radiographic and clinical information. The novel database was developed using a novel, efficient neuroradiology workflow that incorporated a UNETR algorithm into PACS at Yale, allowing segmentation of over 100 gliomas per month. Increasing research in AI application to glioma analysis and treatment is supported at present by publicly available glioma databases. Current prominent examples include the UCSF Preoperative Diffuse Glioma MRI Dataset (n=501)2, The CancerGenome Atlas’ glioblastoma dataset available at The Cancer Imaging Archive (n=262)6, and the Multimodal Brain Tumor Segmentation (BraTS) challenge dataset (n=542)7. Our dataset includes FLAIR and T1 post-contrast images of a larger number of tumors (n=1,033), represents all 4 grades of glioma (595 Grade 4, 96 Grade 3, 105 Grade 2, 45 Grade 1, 192 unknown), and features significant clinical information such as patient characteristics, tumor classification, and molecular subtype.Conclusion
We present a novel dataset consisting of 1,033 glioma patients, including segmented tumors on FLAIR and T1 post-contrast images, additional qualitative imaging features, and clinical information. Open access to this database will provide critically necessary data for the development of novel algorithms to support the analysis and treatment of CNS tumors.Acknowledgements
No acknowledgement found.References
1. Aboian M, Bousabarah K, Kazarian E, Zeevi T, Holler W, Merkaj S, Cassinelli Petersen G, Bahar R, Subramanian H, Sunku P, Schrickel E, Bhawnani J, Zawalich M, Mahajan A, Malhotra A, Payabvash S, Tocino I, Lin M, Westerhoff M. Clinical implementation of artificial intelligence in neuroradiology with development of a novel workflow-efficient picture archiving and communication system-based automated brain tumor segmentation and radiomic feature extraction. Front Neurosci. 2022 Oct 13;16:860208. doi: 10.3389/fnins.2022.860208. PMID: 36312024; PMCID: PMC9606757.
2. Calabrese, Evan, et al. “The University of California San Francisco Preoperative Diffuse Glioma MRI Dataset.” Radiology: Artificial Intelligence, vol. 4, no. 6, 2022, https://doi.org/10.1148/ryai.220058.
3. New ACR DSI Searchable FDA-Cleared Algorithm Catalog Can Ease Medical Imaging AI Integration. Accessed November 9, 2022. https://www.acrdsi.org/News-and-Events/New-ACR-DSI-Searchable-FDA-Cleared-Algorithm-Catalog-Can-Ease-Medical-Imaging-AI-Integration
4. Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21(Suppl 5):v1-v100. doi: 10.1093/neuonc/noz150. PubMed PMID: 31675094; PubMed Central PMCID: PMC6823730.
5. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803-20. doi: 10.1007/s00401-016-1545-1. PubMed PMID: 27157931.
6. Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4(1):170117.
7. Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learningalgorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS Challenge. http://arxiv.org/abs/1811.02629.
Figures