4481
Best Response Constraint Generative Adversarial Network for Diffusion MRI-based Estimation of Cortical micro-Architecture1Department of Radiology, Children's Hospital of Philadelphia, Philadelphia, PA, United States, 2Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, United States, 3Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States, 4School of Information Science and Engineering, Dalian University of Technology, Dalian, China
Synopsis
Keywords: Diffusion/other diffusion imaging techniques, Microstructure, machine learning/artificial intelligence, neuro
Advanced diffusion MRI (dMRI) has enabled noninvasive assessment of conventional cortical histological measures. However, analytical models are limited by their restrictive model assumptions and lack of validation from quantitative histology. We have developed a Diffusion-MRI based Estimation of Cortical micro-Architecture (DECAM) method using a novel deep learning technique Best Response Constraint Generative Adversarial Network (BRC-GAN) for accurately estimating cortical soma density (SD) leveraging rich dMRI data information. By providing high-fidelity, reproducible whole-brain estimated SD maps validated with histology, DECAM paves the way for data-driven noninvasive virtual histology for potential applications such as Alzheimer’s diseases.Purpose
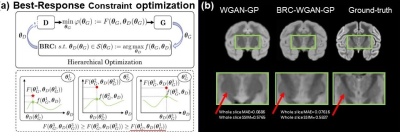
Advanced diffusion MRI (dMRI) has enabled noninvasive assessment of conventional cortical histological measures1-9. However, analytical models are limited by their restrictive model assumptions and lack of validation from quantitative histology. Traditional machine learning methods such as random forest regression lack the rich neighborhood information in diffusion MRI image patches and provide overly simplistic model for representing the complex relationship between dMRI signal and microstructure. By introducing a discriminator network that teaches the generative network how realistic the generative outputs are compared to ground-truth, Generative Adversarial Networks (GANs) can greatly improve the fidelity in image generation and prediction tasks. The Best Response Constraint further improves the GAN by putting a constraint on the discriminator parameters that only allows for the best performing discriminator that optimizes the discriminator cost function10(Fig. 3a). Based on best response constraint (BRC) generative adversarial network (GAN), we have developed Diffusion-MRI based Estimation of Cortical micro-Architecture (DECAM)11, a deep learning and dMRI-based method accurately estimating cortical soma density (SD) leveraging complementary information in diffusion weighted image volumes. By providing high-fidelity and reproducible estimated SD maps validated with histology, DECAM paves the way for data-driven noninvasive virtual histology for potential applications such as Alzheimer’s diseases.Methods
DMRI for macaque (Fig. 1a)DMRI acquisitions with two b-values (b=1500, 4500s/mm2) and 30 gradient directions were performed on a normal postmortem Rhesus macaque brain with in-plane resolution 0.6×0.6mm2, slice thickness=2mm, 2 repetitions for each b-value.
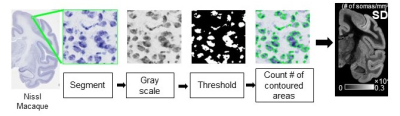
Quantification of soma density from histological images
For measuring SD for macaques (Fig. 2), the Nissl-stained histology images of resolution 0.46µm/pixel12 were blocked into segments with size of 0.24×0.24 mm2, respectively. Segments are gray-scaled and threshold. SD is defined as number of contoured areas/ segment area in mm2. The calculated SD map agrees well with SD from histology13.
Histology-MRI registration (Fig. 1a)
10 Nissl histology slices were gray-scaled and affine registered to average diffusion weighted image (aDWI). SD map was affine registered to aDWI using the same transformation. The registered dMRI volumes and SD maps were cut to 3 by 3 patches and served as training data.
Training Best Response Constraint (BRC) Generative Adversarial Network (GAN)
By introducing a discriminator network that teaches the generative network how realistic the generative outputs are compared to ground-truth, GANs can greatly improve the fidelity in image generation and prediction tasks. The Best Response Constraint further improves the GAN by putting a constraint on the discriminator parameters that only allows for the best performing discriminator that optimizes the discriminator cost function10 (Fig. 3a). More specifically, the BRC is a constraint S(θG) on discriminator parameters θD that maximizes the discriminator cost function f(θG, θD). The generator parameters θG optimize the generator cost function only within the constraint that θD stays in S(θG). We implemented a Wasserstein GAN with gradient penalty (WGAN-GP) with the BRC and trained the model with an Adam optimizer with beta=0.9999 and learning rate=0.0002 for 40 epochs. A mean-squared-error cost function between predicted images and ground-truth was added to the generator cost function to aid prediction.
Estimation of soma density
Whole brain SD was estimated on two separate macaque subjects not used for training the network, and SD was also predicted for a held-out slice on the training macaque brain.
Results
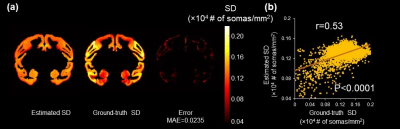
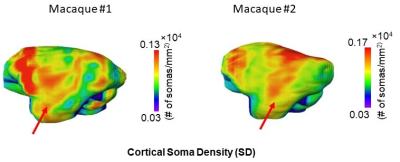
Fig.3b shows the improvement of BRC-WGAN-GP over regular WGAN-GP on a held-out slice for predicting Nissl staining intensity. In the deep gray matter (GM) region, the predicted Nissl staining from BRC-WGAN-GP can clearly show the contrast between deep GM and the internal capsule, while traditional WGAN-GP cannot (red arrows). Estimated SD from noninvasive DECAM on macaque brain are shown in Fig. 4a. A high spatial correspondence and a low residual between histology ground-truth and DECAM-derived SD (Fig. 4a) maps can be observed. Moreover, the estimated SD significantly correlate with the ground-truth values (Fig. 4b) (p<0.0001, Pearson correlation coefficient r=0.53). The mean absolute error (MAE) for predicting SD is 0.0235×104 somas/mm2. Fig. 5 shows reproducible estimated SD on two test macaque subjects not used for training. The pattern of higher soma density in the superior temporal gyrus can be clearly appreciated (red arrows).Discussion and conclusion
We qualitatively and quantitatively demonstrate high correspondence between the DECAM-estimated SD maps and ground-truth in macaques. We also show reproducibility across subjects for whole-brain cortical SD maps. BRC-WGAN-GP outperforms traditional GAN in deep gray matter region important to Alzheimer’s pathology. By providing high-fidelity, reproducible estimated SD and ND maps validated with histology, DECAM paves the way for paradigm-shifting data-driven noninvasive virtual histology for potential applications such as Alzheimer’s diseases.Acknowledgements
This study is funded by NIH R01MH092535, R01MH125333, R01EB031284, R21MH123930 and P50HD105354.References
1. Stanisz GJ, Wright GA, Henkelman RM, Szafer A. An analytical model of restricted diffusion in bovine optic nerve Magn Reson Med. . 1997;37:103-111.
2. Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional kurtosis imaging: the quantification of non‐gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med. 2005;53:1432-1440.
3. Zhou XJ, Gao Q, Abdullah O, Magin RL. Studies of anomalous diffusion in the human brain using fractional order calculus. Magn Reson Med. 2010;63:562-569.
4. Jespersen SN, Kroenke CD, Østergaard L, Ackerman JJ, Yablonskiy DA. Modeling dendrite density from magnetic resonance diffusion measurements. NeuroImage. 2007;34:1473-1486.
5. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage. 2012;61:1000-1016.
6. Le Bihan D, Johansen-Berg H. Diffusion MRI at 25: exploring brain tissue structure and function. NeuroImage. 2012;61:324-341.
7. Alexander DC, Dyrby TB, Nilsson M, Zhang H. Imaging brain microstructure with diffusion MRI: practicality and applications. NMR Biomed. 2019;32:e3841.
8. Novikov DS, Fieremans E, Jespersen SN, Kiselev VG. Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation. NMR Biomed. 2019;32:e3998.
9. Zhu T, Peng Q, Ouyang A, Huang H. Neuroanatomical underpinning of diffusion kurtosis measurements in the cerebral cortex of healthy macaque brains. Magn Reason Med. 2021; 85:1895-1908.
10. Liu, R., Gao, J., Liu, X., & Fan, X. Revisiting GANs by Best-Response Constraint: Perspective, Methodology, and Application. 2022; arXiv preprint arXiv:2205.10146.
11. Zhu T, Ouyang M, Huang H. Diffusion-MRI based Estimation of Cortical Architecture using Machine-learning (DECAM). 2022; ISMRM 2022 abstract number: 0257
12. Mikula S, Trotts I, Stone JM, Jones EG. Internet-enabled high-resolution brain mapping and virtual microscopy. 2007; NeuroImage. ;35:9-15. 13. Carlo CN, Charles FS. Structural uniformity of neocortex, revisited.2013; PNAS. 110.4: 1488-1493.
Figures