4479
Diffusion spectrum imaging at 7T for probing Restricted compartment of Textile-based Phantom1Learning Research and Development Center, University of Pittsburgh, Pittsburgh, PA, United States, 2Department of Developmental Biology, University of Pittsburgh, Pittsburgh, PA, United States, 3Rangos Research Center Animal Imaging Core, University of Pittsburgh, Pittsburgh, PA, United States, 4Psychology Software Tools, Pittsburgh, PA, United States, 5Psychology, University of Pittsburgh, Pittsburgh, PA, United States, 6Bioengineering, University of Pittsburgh, Pittsburgh, PA, United States, 7Neurosurgery, University of Pittsburgh, Pittsburgh, PA, United States, 8Radiology, University of Pittsburgh, Pittsburgh, PA, United States
Synopsis
Keywords: Phantoms, Quantitative Imaging, Diffusion MRI, microstructural imaging
Validating mathematical models that describe the diffusion in the restricted compartment in biological tissue is an active research area. Techniques like NODDI, SMT, and CHARMED models claim to quantify restricted, hindered, and free water compartment but lacks validation on a typical clinical scanner. We present a 3D-printed phantom that uses (non)-water-filled textile-based hollow fiber with a 0.9μ diameter to validate restricted and hindered compartments. This study used a DSI-based scanning protocol and reconstruction method (GDSI and RDI) to probe the restricted compartment at a 2-6μ length scale. Scalar metrics derived from reconstruction methods show a significant difference between water-(un)filled taxons.Introduction
Diffusion MRI-based acquisition protocol and mathematical models are used to quantify restricted and hindered tissue compartments7,9,10,11. These scanning techniques involve q-space sampling, varying b-value, diffusion gradient directions (multi-shell15 and diffusion spectrum imaging14), and reconstructing ensemble average propagator7,15,1 (EAP). Reconstruction methods like CHARMED7, NODDI6, and SMT5 directly model diffusivities and volume fraction of each compartment versus methods like generalized q-sampling imaging1, restricted diffusion imaging2, generalized diffusion spectrum imaging3 and restriction diffusion spectrum imaging16 estimated first EAP, and then it uses to drive markers that correlate to the compartments.Validating these models is very challenging in the biological tissue due to the complexity of white matter tissue9,10,11. To overcome these complexities, we need an idealized, replicable, and scalable phantom11,12,13 that can mimic each compartment with known manufactured fibers and density patterns. Ideally, these synthetic fibers should have a μ-level diameter between 0.5-2μ to mimic the restricted compartment-like white-matter tissue. The hindered compartment is created by controlling the density pattern of these fibers in a cube.
We propose two variants of diffusion-MRI phantoms suitable for scanning on a 7T animal scanner. Each phantom contains a similar cube with taxons with a 0.8μ diameter a) filled with pure water and b) filled with dissolvable copolyester. The first variant mimics restricted (inside fiber) and hindered compartments (outside fiber), and the second mimics only hindered compartments. RDI1,2 and GDSI3-based derived metrics quantify restricted, hindered, and free water compartments. We found a significant difference between water-filled-vs-unfilled taxons in RDI-based metrics that probe compartments at 2μ, 4μ and 6μ length scales and GDSI-based markers that probe at 1μ, 2μ, 4μ, 6μ, and 8μ length scale. Length scale less than 2μ requires a higher b-value, ie. high gradients.
Method
Phantom Construction:Both variants of the phantom are created by 3D printing a cubic shell that is hollow inside to hold the filled and non-filled versions of taxons. Taxsons12,13 is created using nylon and can contain dissolvable copolyester or water. Water filling for the second variant requires four weeks of treatment; see for detail13. Both phantoms are further put inside a cylinder filled with pure water.
Data acquisition:
Both phantoms were scanned on a 7T Bruker animal scanner with bmax=5000 s/mm2 with 107 non b=0 non-colinear gradient directions and 15 b=0 s/mm2 with a total of 122 diffusion-weighted images. Stimulated echo (STE) is used with the BCC sampling scheme of Q-space4. Other MR parameters are as follows; FoV 3.0x3.0cm: voxel size: 0.117x0.117x0.5mm3 with 20 axial slices, TE=12.31ms, TR=5000ms, δ =2ms and Δ=80ms.
Reconstruction: Generalized Diffusion Spectrum Imaging
DSI datasets for both phantoms are reconstructed to recover EAPs using generalized diffusion spectrum imaging3 (GDSI). An in-house PyTorch-based python program is implemented that takes 12 hours of computation time on NVIDIA GPU 2080Ti. Recovered EAPs for each voxel are sampled in 1024 angular directions and 100-sample radially between 0.0 and 1.0 (with 1.0 being the mean displacement distance of free water3). The scalar metric is derived for the estimated EAP, Pr as probability at displacement r averaged across all directions (r=1,2,4,6,8μ; see Fig 2).
Reconstruction: Restricted Spectrum Imaging
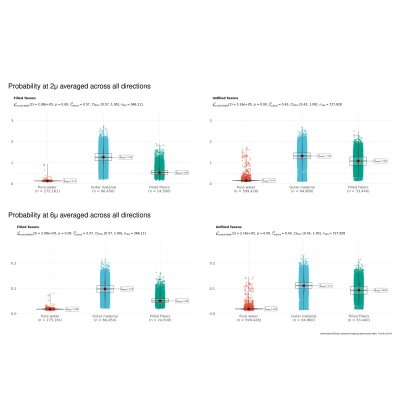
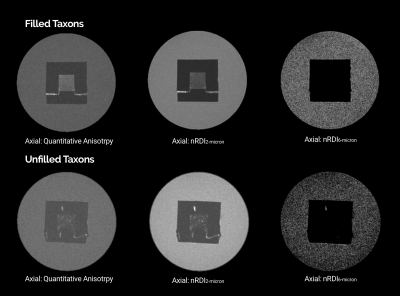
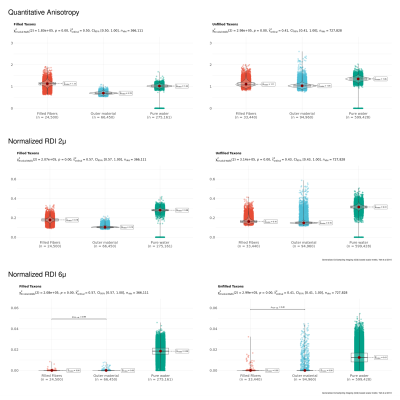
Another reconstruction method, Restricted Spectrum Imaging, estimates non-restricted diffusion imaging (nRDI) for three length scales, L=2,4,6μ, using DSI Studio1,2. In addition, a quantitative anisotropy (QA) map is also estimated using GQI reconstruction algorithms (Fig 4). All derived metrics from GDSI (P1μ, P2μ, P4μ, P6μ, P8μ) and GQI/RDI (QA, nRDI2μ, nRDI4μ, nRDI6μ) are further used for the statistical test for three ROIs (Pure water, Outer 3D material, and water-filled taxons). Statistical tests and box/violin plots (Figures 3 and 5) are created using R's ggplot package.
Results and Discussions
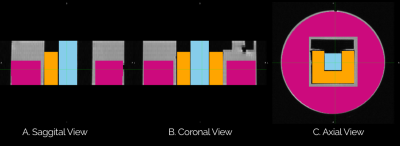
Fig. 2 shows the axial and transversal view of biomarkers estimated using GDSI with five different length scale for filled and non-filled taxons. Sagittal views show a stripped-like pattern for the filled taxons compared to non-filled taxons, indicating the restricted compartment's length scale. Fig. 3 shows the comparison between three ROIs for each GDSI-derived metric. Each metric shows a significant difference between pure water and restricted compartment, and this difference decreases at a larger length scale. Fig. 4 shows the axial and transversal view of RDI-derived quantitative anisotropy and non-restricted diffusion map at 2μ and 6μ-length scales. At 6μ, the restricted compartment disappeared. On the other hand, QA and nRDI2μ show uniform values within the cube with water-filled taxons. Fig. 5 shows the statistical difference between 3-ROIs for each nRDI and QA metric. QA and nRDI2μ show a significant difference between pure water and restricted compartment, and the difference reduces at 6μ.Conclusion
We proposed a design of diffusion-MRI phantom, which is used for probing restricted compartments at different diffusion length scales and suitable for a 7T scanner for a high spatial-resolution scan. We have shown that a DSI-based acquisition protocol and reconstruction methods can quantify restricted and hindered compartments. In addition, this phantom provides an idealized bio-mimicking tissue structure with known diameters suitable for validation of other diffusion methods. Future studies include calibration across scans and creating restricted compartments with multiple diameters.Acknowledgements
- Our work on the fibers, fixtures, and routing of fiber paths is supported by (NIH/NINDS, R44-NS103729)
- This project was funded by the. NIH/NINDS, R44-NS103729, DoD project W81XWH-20-1-0774, Veteran Administration Contract VA I01RX003444 and the David Scaife Foundation of Pittsburgh
References
[1] Yeh, F. C., Wedeen, V. J., & Tseng, W. Y. I. (2010). Generalized q-sampling imaging. IEEE transactions on medical imaging, 29(9), 1626-1635.
[2] Yeh, F. C., Liu, L., Hitchens, T. K., & Wu, Y. L. (2017). Mapping immune cell infiltration using restricted diffusion MRI. Magnetic resonance in medicine, 77(2), 603-612.
[3] Tian, Q., Yang, G., Leuze, C., Rokem, A., Edlow, B. L., & McNab, J. A. (2019). Generalized diffusion spectrum magnetic resonance imaging (GDSI) for model-free reconstruction of the ensemble average propagator. NeuroImage, 189, 497-515.
[4] Chiang, W. Y., Wedeen, V. J., Kuo, L. W., Perng, M. H., & Tseng, W. Y. (2006). Diffusion spectrum imaging using body-center-cubic sampling scheme. In Proc. 14th Annu. Meeting ISMRM (p. 1041).
[5] Kaden E, Kelm ND, Carson RP, Does MD, and Alexander DC: Multi-compartment microscopic diffusion imaging. NeuroImage, vol. 139, pp. 346–359, 2016
[6] Zhang, Hui, et al. "NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain." Neuroimage 61.4 (2012): 1000-1016
[7] Assaf, Yaniv, and Peter J. Basser. "Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain." Neuroimage 27.1 (2005): 48-58
[8] Li, Hua, et al. "Linking spherical mean diffusion weighted signal with intra-axonal volume fraction." Magnetic resonance imaging 57 (2019): 75-82.
[9] Jelescu, I. O., & Budde, M. D. (2017). Design and validation of diffusion MRI models of white matter. Frontiers in physics, 5, 61.
[10] Nilsson, M., van Westen, D., Ståhlberg, F., Sundgren, P. C., & Lätt, J. (2013). The role of tissue microstructure and water exchange in biophysical modelling of diffusion in white matter. Magnetic Resonance Materials in Physics, Biology and Medicine, 26(4), 345-370.
[11] Huang, S. Y., Witzel, T., Keil, B., Scholz, A., Davids, M., Dietz, P., ... & Rosen, B. R. (2021). Connectome 2.0: Developing the next-generation ultra-high gradient strength human MRI scanner for bridging studies of the micro-, meso-and macro-connectome. NeuroImage, 243, 118530.
[12] Fan, Q., Nummenmaa, A., Wichtmann, B., Witzel, T., Mekkaoui, C., Schneider, W., ... & Huang, S. Y. (2018). Validation of diffusion MRI estimates of compartment size and volume fraction in a biomimetic brain phantom using a human MRI scanner with 300 mT/m maximum gradient strength. Neuroimage, 182, 469-478.
[13] Zuccolotto, Anthony P., et al. "Mri phantom including hollow fluid filled tubular textiles for calibrated anisotropic imaging." U.S. Patent Application No. 16/859,444.
[14] Wedeen, V. J., Hagmann, P., Tseng, W. Y. I., Reese, T. G., & Weisskoff, R. M. (2005). Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magnetic resonance in medicine, 54(6), 1377-1386.
[15] Jeurissen, B., Tournier, J. D., Dhollander, T., Connelly, A., & Sijbers, J. (2014). Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage, 103, 411-426.
[16] White, N. S., Leergaard, T. B., D'Arceuil, H., Bjaalie, J. G., & Dale, A. M. (2013). Probing tissue microstructure with restriction spectrum imaging: histological and theoretical validation. Human brain mapping, 34(2), 327-346.
Figures