4460
Influence of Temporal Sampling on Reproducibility of Radiomics Features in Cardiac Cine MRI1Institute for Computer-assisted Cardiovascular Medicine, Charité - Universitätsmedizin Berlin, Berlin, Germany, 2DZHK (German Centre for Cardiovascular Research), Partner Site Berlin, Berlin, Germany, 3Fraunhofer MEVIS, Bremen, Germany, 4Charité - Universitätsmedizin Berlin, Berlin, Germany, 5Deutsches Herzzentrum Berlin, Berlin, Germany, 6University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Synopsis
Keywords: Heart, Radiomics, Reproducibility
Radiomics has been applied in cardiovascular imaging with promising results. Still, the effect of imaging parameters on the reproducibility of radiomics features needs to be well understood for these to be used in clinical diagnostics. We conduct a retrospective study on short-axis cine CMR images of healthy volunteers to assess the impact of different temporal sampling parameters on the reproducibility of shape, first-order, texture, and deformation field-based features, and contextualize the findings with inter- and intra-observer variability. We find that the reproducibility of features is dependent on temporal sampling, warranting the introduction of parameter standardization for radiomics analyses.Introduction
Radiomics is a technique to quantify intensity patterns and shape features of anatomical structures in medical image data which could help identify pathological changes. Applied to cardiac imaging, radiomics has shown to assist with phenotyping cardiac diseases, e.g. for the assessment of atherosclerosis or myocardial scarring1-4. As radiomics features are calculated on intensity values in a pixel or voxel grid, differences in imaging parameters and preprocessing typically chosen to optimize visual image interpretation are likely to affect the feature values. In cine MRI, the choice of temporal sampling rate has been shown to impact the measurement of functional parameters including ventricular volumes and myocardial strain5-8.While the “Image Biomarker Standardization Initiative” (IBSI) has been working towards establishing standardized definitions of radiomics features and their implementation9, no standard for imaging parameters and preprocessing for cardiac radiomics currently exists, compromising the quality of studies employing radiomics in CMR10. Previous studies on the reproducibility of radiomics features do not employ consistent definitions of reproducibility and use varying thresholds to interpret Intraclass Correlation Coefficients (ICC). In this study we first establish the inter- and intra-observer variability of the image segmentations before investigating the effect of different temporal sampling rates on the reproducibility of radiomics and cardio-specific features calculated for cine MRI of healthy volunteers.
Methods
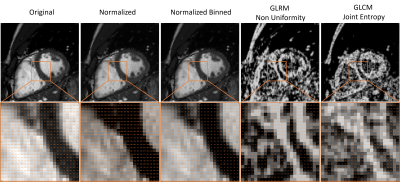
In this retrospective study, two experts independently segmented the left ventricular myocardium (LVM), left ventricular blood pool (LVBP) and right ventricular blood pool (RVBP) in cine CMR images of eight healthy volunteers (50% female), twice at high temporal resolution of 50 frames per cardiac cycle (fpc), and one of the observers segmented at additional resolutions of 20, 30 and 40fpc once (all short-axis ECG-triggered bSSFP cine CMR, 5mm slice thickness, TE/TR/flip=1.3-2.1ms/2.6-4.1ms/60º, Achieva 1.5T, Philips Healthcare, Best, NL). Preprocessing consisted of intensity rescaling to DICOM standard range [0, 4095] and discretization with bin width 25 (PyRadiomics default11). The observers reviewed and refined the spline-based contours proposed by a neural network in a semi-automatic segmentation tool12, before the contour was rasterized for further processing.The inter- and intra-observer variability of segmentation was measured with the Dice Score (DSC), Hausdorff Distance (HD) and Average Contour Distance (ACD).
Three categories of features were calculated:
- three 2D shape features representing standard area/volume assessment plus sphericity and elongation (for each of LVM, LVBP, RVBP),
- 15 first-order and texture features identified as reproducible in the literature (for each of LVM, LVBP, RVBP),
- six features describing cardiac anatomy and motion, including strain features calculated on deformation fields obtained from quadrature filtering and a measure of fractal dimension13.
Results
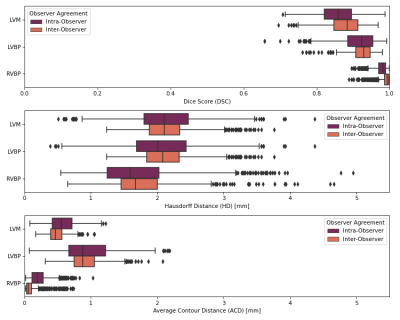
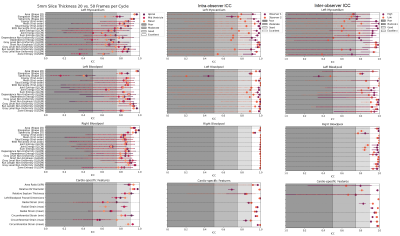
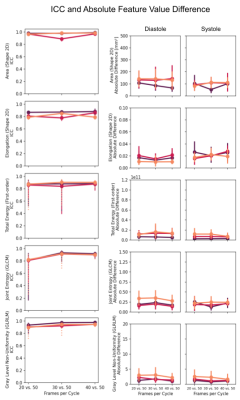
The inter- and intra-observer metrics are: mean DC ≥0.88, mean HD ≤2.3mm and mean ACD ≤1.0 mm (Figure 2).The radiomics and cardiac-specific feature ICC values are displayed in Figure 3 for the extremum case (20 vs 50 fpc) and compared to inter- and intra-observer ICC. The Area is consistently “excellent”, whereas LVM Elongation and LVBP Sphericity are found “good” / ”moderate” and “poor” respectively. Texture-based features are in general less reproducible in the LVM (“moderate” or “good”), whereas they are mostly “good” in the LVBP and RVBP. Cardiac-specific features are found to be “moderate” to “excellent”. With decreasing difference between temporal sampling rates the reproducibility of most features improves. Notable exceptions are the LVM Elongation of the basal slice, the LVBP Sphericity in all slices, and the maximum Radial and Circumferential Strain in the basal and mid-ventricular slices, where the higher similarity in temporal sampling parameters does not lead to higher ICC values. Selected example ICC values and absolute differences across the resolutions are displayed in Figure 4.
Conclusion
The examined shape features depend on the contour of the segmented structure rather than the intensity values of voxels within. We observe high reproducibility of Area across temporal resolutions, suggesting that edges and boundaries are sufficiently preserved at lower temporal resolution to produce consistent segmentations – in fact, the lower inter- and intra-observer consistencies show that observers have comparable effect. Despite the consistency of the LVBP Area, the reproducibility of the Sphericity of the highly circular LVBP is poor across the resolutions, indicating the sensitivity of this feature to small changes in the segmentation contours.First-order and texture features are calculated on the histogram of intensity values in the segmented region. The sub-optimal reproducibility observed for these features suggests that the difference in intensity distribution at different sampling rates cannot be compensated by subsequent resampling. As expected, Radial and Circumferential Strain are dependent on the temporal sampling parameters, but at a comparable scale to standard radiomics features. In the context of the excellent inter/intra-observer reproducibility observed, this study highlights the need for standardization of temporal parameters to be used for radiomics analyses to ensure the reliability of features for clinical use.
Acknowledgements
This work was supported by the German Ministry for Education and Research (BMBF) as BIFOLD - Berlin Institute for the Foundations of Learning and Data (01IS18025A and 01IS18037A), as well as by the German Research Foundation (DFG) as part of SFB-1470, B06.References
1. Kolossvary, M., et al., Radiomic Features Are Superior to Conventional Quantitative Computed Tomographic Metrics to Identify Coronary Plaques With Napkin-Ring Sign. Circ Cardiovasc Imaging, 2017. 10(12).2. Li, X.N., et al., Identification of pathology-confirmed vulnerable atherosclerotic lesions by coronary computed tomography angiography using radiomics analysis. Eur Radiol, 2022. 32(6): p. 4003-4013.
3. Mancio, J., et al., Machine learning phenotyping of scarred myocardium from cine in hypertrophic cardiomyopathy. Eur Heart J Cardiovasc Imaging, 2022. 23(4): p. 532-542.
4. Fahmy, A.S., et al., Radiomics and deep learning for myocardial scar screening in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson, 2022. 24(1): p. 40.
5. Hassani, C., et al., Myocardial Radiomics in Cardiac MRI. AJR Am J Roentgenol, 2020. 214(3): p. 536-545.
6. Mannil, M., et al., Artificial Intelligence and Texture Analysis in Cardiac Imaging. Curr Cardiol Rep, 2020. 22(11): p. 131.
7. Inoue, Y., et al., Effect of temporal resolution on the estimation of left ventricular function by cardiac MR imaging. Magn Reson Imaging, 2005. 23(5): p. 641-5.
8. Backhaus, S.J., et al., Defining the optimal temporal and spatial resolution for cardiovascular magnetic resonance imaging feature tracking. J Cardiovasc Magn Reson, 2021. 23(1): p. 60.
9. Zwanenburg, A., et al., The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology, 2020. 295(2): p. 328-338.
10. Chang, S., et al., Quality of science and reporting for radiomics in cardiac magnetic resonance imaging studies: a systematic review. Eur Radiol, 2022. 32(7): p. 4361-4373.
11. van Griethuysen, et al., Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research, 2017. 77(21), e104–e107.
12. Hüllebrand, M., et al., A Collaborative Approach for the Development and Application of Machine Learning Solutions for CMR-Based Cardiac Disease Classification. Front Cardiovasc Med, 2022. 9: p. 829512.
13. Tautz, L., et al., Cardiac radiomics: an interactive approach for 4D data exploration. Current Directions in Biomedical Engineering, 2020. 6(1).
14. Koo, T.K. and M.Y. Li, A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med, 2016. 15(2): p. 155-63.
Figures