4377
Quantitative MRI T2mapping in the knee joint using deep learning-based reconstruction for Compressed sensing1The First Hospital of Jilin University, Changchun, China, chang chun, China, 2Philips Healthcare, Beijing, China, Beijing, China, China
Synopsis
Keywords: Osteoarthritis, Machine Learning/Artificial Intelligence
MRI T2 mapping has been recommended as a noninvasive biomarker of knee cartilage lesions. However, due to the long acquisition time, it hasn’t been widely used in the clinical setting. Recently, deep learning-based acceleration of compressed sensing (CS) has shown promising results without losing image quality. The purpose of this study was to explore the feasibility of quantitative knee T2-mapping accelerated by deep learning-based compressed sensing (CS-AI), and compare the image quality and diagnostic performance with conventional CS. The results demonstrates that quantitative knee T2 mapping with reconstruction by CS-AI was feasible, suggesting better diagnostic performance without extra time consuming.Introduction
Clinically, the knee joint is the most common site of osteoarthritis(OA)[1].OA has been seen as a clinical and pathological outcome of a series of diseases that can lead to articular cartilage degradation, and eventually functional incapacitation[1, 2]. Due to the excellent high tissue resolution,muti-dimensional imaging and absence of radiation , Magnetic resonance imaging (MRI) is commonly used to study knee osteoarthritis. In addition to good visualization of soft tissue structures, it can also quantitatively detect cartilage changes in the early or possible reverse stages to intervene in disease progression [3, 4]. Currently, quantitative analysis of the T2 mapping musculoskeletal system has attracted widespread interest as a non-invasive biomarker of cartilage and meniscus components[5]. Quantitative T2 mapping has been suggested to provide information on extracellular matrix water content and collagen fiber structure [6]. However, the long scanning time required for this technique prevents its spread in clinical settings. Compressed sensing is a technology that can reduce acquisition time by using an iterative reconstruction algorithm to reduce the number of acquired lines in K-space and restore the missing data[7]. Recently, the application of artificial intelligence to CS leads to better results. For instance, Adaptive-CS-Net, proposed by Pezzotti et al , uses CNN instead of wavelet transform as sparse transform in compressed sensing,and ensures data consistency and domain-specific knowledge[8]. In this study, combination CNN-based sparsifying approach with the image reconstruction approach based on compressed sensing, is presented as Compressed SENSE AI (CS-AI). The purpose of the study is to acquire the quantitative T2 mapping accelerated by CS-AI with different echo times and compare the image quality and diagnosis performance with CS.Methods
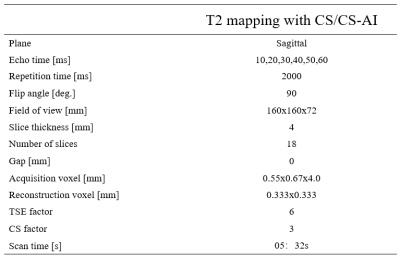
Thirty healthy volunteers were enrolled with informed consent written in this IRB approved study. All examinations were performed on a 3.0T MR system (Ingenia Elition, Philips Healthcare) with a 8-channel receive knee coil. The knee was fixated within the coil to reduce motion artifacts. All the volunteers underwent quantitative T2-mapping using a spin-echo(SE) multi-slice multi-echo(MSME) pulse sequence accelerated by CS-SENSE(CS)and CS-SENSE AI(CS-AI)with six echo times. More details of the sequence parameters are shown in Table1.Image quality was evaluated both subjectively and objectively. The five-point Likert scale was used to assess individual anatomical structures subjectively (1=poor,2=below average,3=fair,4=good,5=excellent). Objective evaluation including Signal-to-Noise Ratio (SNR) and contrast-to-noise ratio (CNR) was performed separately by two radiologists (less than 5 years of experience) and reviewed by a senior radiologist (more than 5 years of experience). Quantitative T2 mapping values were obtained by placing regions of interest (ROI) at different anatomical,which included bone (diatal femur),muscle (gastrocnemius), anterior cruciate ligament(ACL) ,posterior cruciate ligament (PCL) ,cartilage (patella cartilage、femoral cartilage ),joint fluid and adipose tissue. The statistical analysis was performed with SPSS,version 25.0(IBM). The Paired T test was used to evaluate the SNR and CNR. Interobserver correlation and intersequence correlation of detected abnormalities were determined using Cohen’s kappa and Cronbach’s alpha. We divided the P values into three groups: p≤0.05,p≤0.005,p≤0.001.
Results
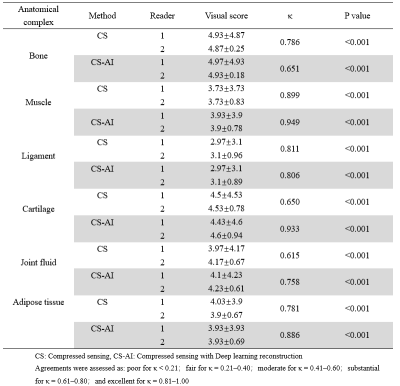
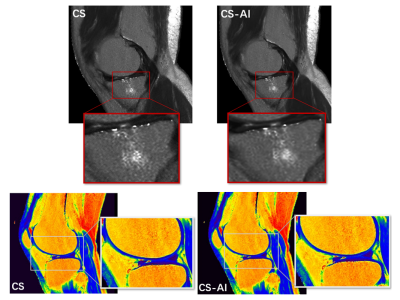
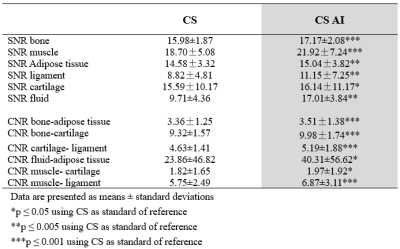
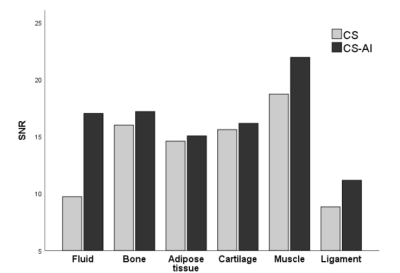
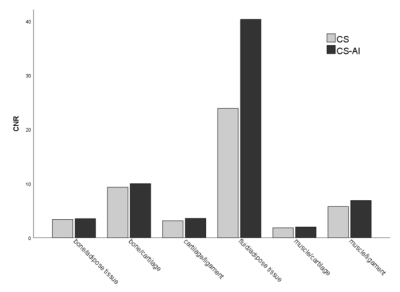
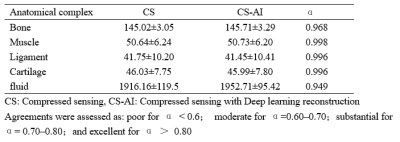
A total of 30 images were evaluated by two radiologists according to Likert scores, as shown in Table 2. It can be seen that the image quality score of CS-AI is higher than that of CS. Figure 1 shows the image quality with and without AI,respectively. Meanwhile, the CS-AI sequence can produce visually sharper images compared to CS. As illustrated in Table 3, SNR and CNR were higher for all tissues in the CS-AI sequence compared to CS only. Compared to CS,SNR in the CS-AI protocol had the most significant differences for bone(15.98±1.87vs.17.17±2.08, p≤0.001; Figure2 and Figure3) and muscle(18.70±5.08 vs. 21.92±7.24, p≤0.001; Figure2 and Figure3) .The intraclass correlation (ICC) coefficient between the CS-AI and CS for cartilage was 0.996, illustrated in Table 4.Discussion
Quantitative knee T2 mapping accelerated by deep learning-based compressed sensing can ensure image quality and have high denoising performance. Compared with conventional CS, it can better detect some small structures, especially knee cartilage, , and can detect early lesions earlier, which plays a good guiding role in clinical practice. Compared with previous studies, the significance of our study lies in the application of a new acceleration technique (CS-SENSE AI ) to the quantitative T2 mapping of the knee joint to early identify small structural lesions such as cartilage.Conclusion
This study demonstrated the feasibility of deep learning-based reconstruction for CST2 mapping in the knee joint, suggesting better diagnostic performance without extra time consuming, is expected to be applied in clinical work, especially in the early identification of knee osteoarthritis.Acknowledgements
No acknowledgement found.References
1. Hunter, D.J. and S. Bierma-Zeinstra, Osteoarthritis. The Lancet, 2019. 393(10182): p. 1745-1759. 2. Bijlsma, J.W.J., F. Berenbaum, and F.P.J.G. Lafeber, Osteoarthritis: an update with relevance for clinical practice. The Lancet, 2011. 377(9783): p. 2115-2126.
3. Atkinson, H.F., et al., MRI T2 and T1rho relaxation in patients at risk for knee osteoarthritis: a systematic review and meta-analysis. BMC Musculoskelet Disord, 2019. 20(1): p. 182.
4. Roemer, F.W., et al., State of the Art: Imaging of Osteoarthritis-Revisited 2020. Radiology, 2020. 296(1): p. 5-21.
5. Roux, M., et al., MRI T2 Mapping of the Knee Providing Synthetic Morphologic Images: Comparison to Conventional Turbo Spin-Echo MRI. Radiology, 2019. 293(3): p. 620-630.
6. Nieminen, M.T., et al., T2 relaxation reveals spatial collagen architecture in articular cartilage: a comparative quantitative MRI and polarized light microscopic study. Magn Reson Med, 2001. 46(3): p. 487-93.
7. Lustig, M., D. Donoho, and J.M. Pauly, Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med, 2007. 58(6): p. 1182-95.
8. Pezzotti, N., et al., An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction. IEEE Access, 2020. 8: p. 204825-204838.
Figures