4359
Cycle Inverse Consistent Deformable Medical Image Registration with Transfomer1Columbia University, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain
Deformable image registration is a crucial part of the medical image process that seeks to estimate an optimal spatial transformation to align two images. Traditional methods neglect the inverse-consistent property and topology preservation of the transformation, which can lead to errors in registration results. To address these issues, we propose the cycle inverse consistent transformer-based deformable medical image registration model. We adopt Swin-UNet to achieve higher registration performance and comprehensively consider the inverse consistency loss function to guarantee a more accurate registration. Our pipeline can be trained to create study-specific templates of images for diagnostic and/or educational purposes.Introduction
Deformable image registration offers unique properties including topology preservation and invertibility of the transformation and can be implemented through deep learning3. One property of image transformations that has recently been exploited for registration is inverse consistency.8 In addition, a transformer-based registration model uses the Swin Transformer as the encoder to capture the spatial correspondence and a CNN decoder to process the information into a dense displacement field.1 We propose a novel medical image registration model that seeks to ameliorate current limitations by combining successful features of previous models: an inverse-consistent, transformer-based, deformable registration model.Method
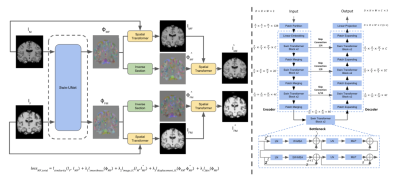
We propose a novel model, as shown in Fig. 1, for medical image registration that seeks to ameliorate the limitations detailed in Fu et al by combining successful features of previous models: an inverse-consistent, transformer-based, deformable registration model that utilizes a Swin-UNet to compute two displacement fields from a pair of input images. After generating the displacement fields, a differentiable spatial transformation network (STN) is applied to generate two warped images based on input images and the displacement fields respectively. Furthermore, the displacement fields are also passed through an inverse module to estimate the inverse displacement fields, and the inverse displacement fields will then be implemented on the warped images to generate the inverse warped image.We propose a multicomponent loss function composed of four components: similarity, smoothness, inverse consistency, and a non-negative Jacobian determinant. We use the structural similarity index measure (SSIM) as the similarity metric to contrast the warped image and the corresponding fixed image. A smoothness constraint on the displacement field is utilized to enforce that be smooth and physically realistic. In addition, since the inverse warped image and the moving image are expected to be similar, the inverse displacement field and displacement field should also be approximately the same; we propose two inverse-consistent loss functions at the image-level and displacement field-level. We also propose the non-negative Jacobian determinant loss to preserve the topology of the warped medical image by penalizing the negative Jacobian determinant that leads to the folding.
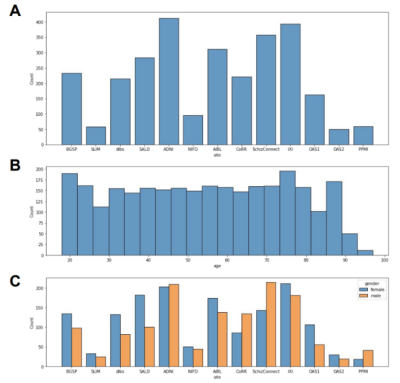
The neuroimaging data10 used in this study were collected from 13 different sites with a total of 2852 T1-weighted brain MRI scans. Our dataset has approximately the same number of scans from different genders and is uniformly distributed over the life span (18 - 97 years old). The dataset is further split into 2282: 285: 285 (8: 1: 1) volumes for train, validation, and test sets, respectively. In our data pre-processing pipeline, we first corrected the bias field inhomogeneity with the N4 bias field correction procedure and affine-registered the raw whole-head scans to the MNI152 unbiased template4,5. Subsequently, the Brain Extraction Tool6 was applied to achieve skull-stripping. After that, we affine-registered these scans to the MNI152 unbiased template.
Results
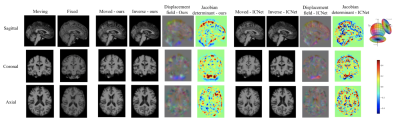
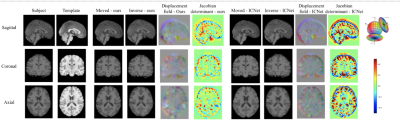
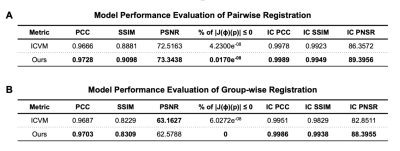
Fig. 5(A) shows the performance of the proposed model and ICNet when dealing with pairwise registration issues, and Fig. 5(B) illustrates the group-wise registration performance of both models. Fig. 3 and Fig. 4 give more details of the registration result by taking one example from each comparison task.Discussion
According to the results shown in Fig. 5, our model benefits from adding inverse consistency loss and achieves higher performance in terms of average PCC, SSIM, and PSNR, indicating a higher similarity between the registered images. Our model has fewer voxels with negative Jacobian determinant value, which means the displacement field is smoother, and the physical appearance of the scans is more realistic upon registration. These assertions are verified by Fig.3 and Fig. 4, in which the results indicate that our model has the capability to preserve the inverse consistency of the transformation by enforcing local refinement, while simultaneously producing registered images with higher similarity. These demonstrated results confirm that adding inverse consistency loss and non-negative Jacobian loss, together with the Swin transformer, has created a model which can learn to perform more physically realistic and accurate registrations than have been reported in the past.Acknowledgements
No acknowledgement found.References
1. Zhu, Y., Lu, S. (2022). Swin-VoxelMorph: A Symmetric Unsupervised Learning Model for Deformable Medical Image Registration Using Swin Transformer. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13436. Springer, Cham. https://doi.org/10.1007/978-3-031-16446-0_8
2. Fu, Y., Lei, Y., Wang, T., Curran, W. J., Liu, T., & Yang, X. (2020). Deep Learning in Medical Image Registration: A Review. Physics in Medicine & Biology, 65(20). https://doi.org/10.1088/1361-6560/ab843e
3. Mok, T. C. W., & Chung, A. C. S. (2020). Fast symmetric diffeomorphic image registration with Convolutional Neural Networks. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). https://doi.org/10.1109/cvpr42600.2020.00470
4. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001 Jun;5(2):143-56. doi: 10.1016/s1361-8415(01)00036-6. PMID: 11516708.
5. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002 Oct;17(2):825-41. doi: 10.1016/s1053-8119(02)91132-8. PMID: 12377157.
6. Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002 Nov;17(3):143-55. doi: 10.1002/hbm.10062. PMID: 12391568; PMCID: PMC6871816.
7. Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., … Chintala, S. (2019). PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32 (pp. 8024–8035). Curran Associates, Inc. Retrieved from http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf
8. Chen, Y., & Ye, X. (2010). Inverse consistent deformable image registration. The Legacy of Alladi Ramakrishnan in the Mathematical Sciences, 419–440. https://doi.org/10.1007/978-1-4419-6263-8_26
9. Balakrishnan, Guha & Zhao, Amy & Sabuncu, Mert & Guttag, John & Dalca, Adrian. (2019). VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Transactions on Medical Imaging. PP. 1-1. 10.1109/TMI.2019.2897538.
10. Feng X, Lipton ZC, Yang J, et al. Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiol Aging. 2020;91:15-25. doi:10.1016/j.neurobiolaging.2020.02.009
Figures