4358
Comparison of Combinations of MTR, FLAIR, and T1WCE MRI Images for Automatic Segmentation of Brain Metastases1Toronto Metropolitan University, Fremont, CA, United States, 2Toronto Metropolitan University, Toronto, ON, Canada, 3Electrical, Computer, and Biomedical Engineering, Toronto Metropolitan University, Toronto, ON, Canada
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Magnetization transfer, Multi-Modal
This study presents an evaluation of utilizing MTR and FLAIR as compared to T1wCE and FLAIR images for automatic segmentation of brain metastases, due to the unknown consequences posed by accumulated contrast enhancement. Numerous combinations of FLAIR, T1wCE, and MTR MRI images were tested on three top-performing segmentation models (MS-FCN, U-NET, and SegresNet) to evaluate the feasibility of using MTR and FLAIR images for tumor segmentation. Overall, the U-Net produced the best similarity scores of 0.4±0.15 using MTR and FLAIR images. A future study will test the utility of 3D MTR images for tumor segmentation using convolutional and full-connected models.Introduction
Diagnosis and radiotherapy treatment planning for patients with brain metastases is currently performed with gadolinium-enhanced contrast-enhanced T1-weighted MRI (T1wCE) and T2-weighted-fluid-attenuated-inversion-recovery (FLAIR) as a reference 1. However, recent evidence suggests that gadolinium (Gd) accumulates in healthy brain tissue and poses unknown consequences for the patient-health, motivating research toward Gd-free treatment planning options 2-5. An alternative to T1wCE brain scans is the evaluation of Magnetization Transfer (MTR) images along with FLAIR modalities for tumor localization in glioblastoma patients 6. In this study, we evaluate the feasibility of MTR images to replace Gd-contrast enhanced images to address the need for Gd-free treatment planning by testing three top-performing Machine Learning tumor segmentation architectures: Multi-scale fully convolutional network (MS-FCN), U-NET, and SegResNet.Methods
The Sunnybrook Hospital-affiliated MRI dataset contained >72 patients’ MTR, FLAIR, and T1wCE images, and the corresponding radiologist-annotated segmentation masks. All images underwent skull stripping to improve the efficacy of image analysis 7. The skull of T1WGd and FLAIR images were removed using the FMIRB Software Library (FSL) brain extraction tool (BET) (version 2.1) 8. The MTR images were skull stripped using HD-BET [29], due to a specific problem where lesions near the skull were removed when FSL was used. The images were then registered using multi-modal geometric registration in MATLAB. Each image was normalised between 0 to 1 to ensure comparability across the dataset using NumPy. The dataset of 72 patients was split into 50 training images, 10 validation images, and 12 testing images, approximately a 70%, 14%, and 16% split, respectively. Each model was evaluated against four different combinations of modalities: FLAIR and T1wCE, MTR and T1wCE, FLAIR and MTR, FLAIR and T1wCE and MTR; 6-fold cross-validation was used to separate training, validation, and test subjects for predictions. Additionally, a binary cross entropy loss function was used. All networks were implemented in Keras (version 2.8.0) 9 underwent individual tuning and were trained for a maximum of 300 epochs, and utilized a learning rate scheduler with a gamma of rate of ⅓ as well as early stopping. Each model was trained using the Adam optimizer, which used momentum and adaptive learning to enable the model to converge faster, and trained with an initial learning rate of 0.001.Results
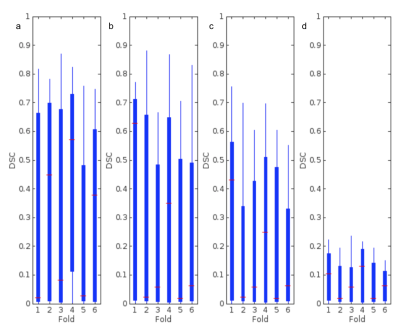
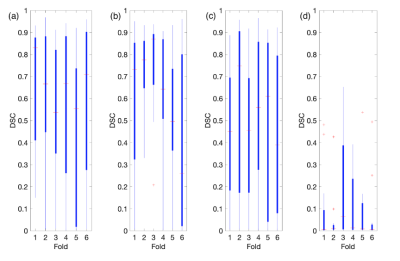
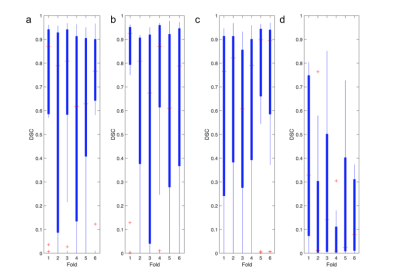
Overall, each model better segmented the tumorous regions from FLAIR and T1wCE modalities over FLAIR and MTR modalities. For T1wCE and FLAIR segmentation, the best performing models were U-Net and FCN, which resulted in average dice similarity coefficient scores (DSC) of 0.70±0.15 and 0.56±0.15 respectively. The best-resulting model amongst the comparable approaches for the FLAIR and MTR sequence is the U-Net which achieved a DSC average of 0.37±0.15, with the FCN averaging a dice score of 0.10±0.15. However, the results achieved in this study show hopeful results for future testing of convolutional and full-connected models to replace T1wCE imaging for tumor segmentation in the future.Discussion
The multiscale fully convolutional network (FCN) used is adapted from a 3D multi-scale FCN utilised by Sun et al. (2020) 6, which achieved state-of-the-art results on the BraTS dataset. This model extracts features along four different pathways and image scales without any fully connected layers, resulting in decreased inference time per image since there are fewer model weights. The U-NET 10 architecture is fully convolutional, utilizing skip connections to preserve all image features after extraction. SegResNet 11. is a recent model utilizing group normalization and block analysis to accurately segment tumorous pixels. In this work, only 2D (256x256) data is used, decreasing the computational demand as well as increasing training and inference speed. Overall, the U-NET architecture was the best-performing segmentation model for FLAIR and MTR images.Conclusion
Overall, each model was able to more accurately segment the tumorous regions from FLAIR and T1wCE modalities over FLAIR and MTR modalities. For T1wCE and FLAIR segmentation, the best performing models were U-Net and FCN, which resulted in average dice similarity coefficient scores (DSC) of 0.70±0.15 and 0.56±0.15 respectively. The best-resulting model amongst the comparable approaches for the FLAIR and MTR sequence is the U-Net which achieved a DSC average of 0.37±0.15, with the FCN averaging a dice score of 0.10±0.15. Overall, while tumor segmentation accuracy is higher from T1wCE and FLAIR images, the results indicate there is potential for use of MTR images. A future, a larger study will test the utility of 3D MTR images for tumor segmentation using convolutional and full-connected models.Acknowledgements
No acknowledgement found.References
1. R. Singh et al., ‘Epidemiology of synchronous brain metastases’, Neuro-Oncology Advances, vol. 2, no. 1, p. vdaa041, Jan. 2020, doi: 10.1093/noajnl/vdaa041.
2. R. J. McDonald et al., ‘Comparison of Gadolinium Concentrations within Multiple Rat Organs after Intravenous Administration of Linear versus Macrocyclic Gadolinium Chelates’, Radiology, vol. 285, no. 2, pp. 536–545, Nov. 2017, doi: 10.1148/radiol.2017161594.
3. A. Kiviniemi, M. Gardberg, P. Ek, J. Frantzén, J. Bobacka, and H. Minn, ‘Gadolinium retention in gliomas and adjacent normal brain tissue: association with tumor contrast enhancement and linear/macrocyclic agents’, Neuroradiology, vol. 61, no. 5, pp. 535–544, May 2019, doi: 10.1007/s00234-019-02172-6.
4. T. Kanda et al., ‘Gadolinium-based Contrast Agent Accumulates in the Brain Even in Subjects without Severe Renal Dysfunction: Evaluation of Autopsy Brain Specimens with Inductively Coupled Plasma Mass Spectroscopy’, Radiology, vol. 276, no. 1, pp. 228–232, Jul. 2015, doi: 10.1148/radiol.2015142690.
5. R. J. McDonald et al., ‘Intracranial Gadolinium Deposition after Contrast-enhanced MR Imaging’, Radiology, vol. 275, no. 3, pp. 772–782, Jun. 2015, doi: 10.1148/radiol.15150025.
6. J. Sun, Y. Peng, Y. Guo, and D. Li, ‘Segmentation of the multimodal brain tumor image used the multi-pathway architecture method based on 3D FCN’, Neurocomputing, vol. 423, pp. 34–45, Jan. 2021, doi: 10.1016/j.neucom.2020.10.031.
7. S. M. Smith, ‘Fast robust automated brain extraction’, Hum Brain Mapp, vol. 17, no. 3, pp. 143–155, Nov. 2002, doi: 10.1002/hbm.10062.
8. F. Isensee et al., ‘Automated brain extraction of multisequence MRI using artificial neural networks’, Hum Brain Mapp, vol. 40, no. 17, pp. 4952–4964, Dec. 2019, doi: 10.1002/hbm.24750.
9. F. Chollet et al., ‘Keras’, 2015. Available: https://github.com/fchollet/keras.
10. O. Ronneberger, P. Fischer, and T. Brox, ‘U-Net: Convolutional Networks for Biomedical Image Segmentation’, arXiv:1505.04597 [cs], May 2015, Accessed: Mar. 20, 2022. [Online]. Available: http://arxiv.org/abs/1505.04597
11. Gao, S., Zhang, S., Liang, K., Liu, Y., Zhang, Z., Li, Z., Yin, Z.: Solution of team. SEALS for ISLES22. In: International MICCAI Brain lesion Workshop. Deepwise AILab and Beijing University of Telecommunication, Beijing, China (2022)
Figures
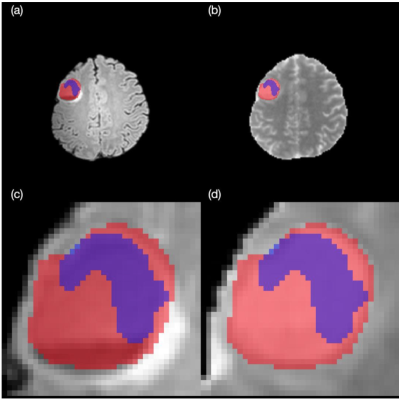
FLAIR-MTR segmentation of P127 from MS-FCN architecture, (a) FLAIR whole brain, (b) MTR whole brain, (c) FLAIR lesion, (d) MTR lesion.