4357
Breast Tumor Segmentation using U-Net with ResNet341BME, UNIST, Ulsan, Korea, Republic of, 2Nuclear Engineering, UNIST, Ulsan, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Animals
The proposed research implemented an automatic tumor segmentation application on an orthotopic breast tumor model. This application can segment the tumors accurately and monitor tumor growth and the therapeutic effect of Doxorubicin for treatment. Also, the outputs from the application can reconstruct into 3D rendering and offer the visualization of shape and volume. As a result, the application can be applied to orthotopic breast tumor model research.Introduction
Breast cancer is the most common cancer in the world, especially the first leading cause of cancer death among women. The major cause of death is metastasis, which cannot be fully mimicked in vitro because of its complexity1. Therefore, orthotopic breast tumor model is significant to the researcher, which shares many features of human primary tumor growth and metastasis.However, there are limitations in traditional orthotopic model research. First, when the researcher measures the tumor volume, a vernier caliper is usually used on skin and it is an inaccurate tumor measuring method. It also leads to another limitation that the method cannot consider therapeutic efficacy for treatment, which can affect the volume or shape of tumors. Also, a major limitation is detecting tumors from MR images. The manual expert can judge tumors after monitoring continuous MR slices, which is a time-consuming task, and if the data size is limited2, identifying small tumors are may difficult.
Therefore, there is a necessity for automatic segmentation. The application can offer fast and accurate segmentation, detect small tumors on a single MR image, automatic evaluation of therapeutic effect, and apply to orthotopic mouse model research. Consequently, the purpose of our study is to generate an automatic segmentation application using deep learning, that can be used to track tumor growth and monitor therapeutic effect on untreated and treated group with Doxorubicin.
Materials and Methods
Data acquisitionTo generate the orthoptic breast tumor model, the MDA-MB-231 cell line was cultured and injected into the mammary fat pad of a nude mouse. The orthotopic breast tumor model was divided into untreated and treated groups with 0.1mg/20g of Doxorubicin. T2-weighted (T2W) data was obtained by fast spin echo (RARE) sequence every 5 days. The MR parametersp; TE, TR, matrix size, FOV, and slice thickness was set to 35ms, 4000ms, 256*256*50, 35*35*25, and 0.5 respectively. The binary tumor masks were created by a manual expert. 1979 MR slices are used as the trainset, the 1st testset consisted of 531 MR slices of untreated groups, and 2nd testset consisted of 833 MR slices of treated groups.
Train and test
The proposed research utilized U-Net3 with ResNet344. U-Net is a network specialized in medical image segmentation. To improve the performance of segmentation, the encoder of U-Net was replaced by pre-trained ResNet34. The network was trained with 5-fold cross-validation and various augmentation techniques were applied during training to avoid overfitting. Also, Dice and Intersection over Union (IoU) were used for evaluation and the training parameters; epoch, batch size, and learning rate, activation was set to 500, 16, and 0.0004, Sigmoid respectively.
Results
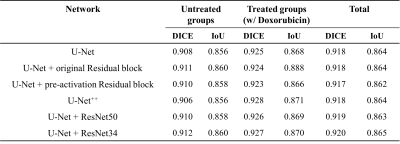
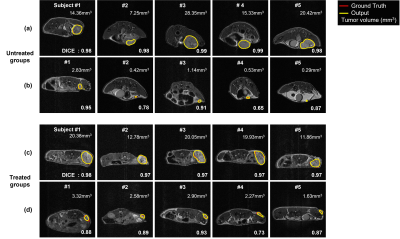
The tumor segmentation performance of various network architectures is shown in Table 1. The U-Net with ResNet34 showed the highest segmentation performance with DICE of 0.92 among the architectures because this architecture extracted the features from the inputs by replacing the encoder part.Figure 2(a) is the test outputs with the best DICE of the untreated groups. Figure 2(b) is the test outputs with the small tumor volume of the untreated groups. Figure 2(c) is also the test outputs with the best DICE of the treated groups. Figure 2(d) is the test outputs with the small tumor volume of the treated groups. The range of small tumor volume is below 5mm3.
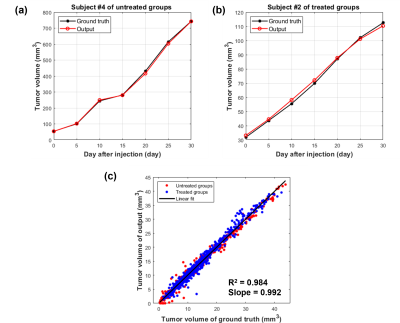
In figure 3(a) and (b), the tumor volume growth of outputs from the trained model was compared to ground truth. Figure 3(a) is subject 4, the biggest tumor volume among the untreated groups, and figure 3(b) is subject 2, the smallest tumor volume among the treated groups. Figure 3(c) is the scatter plot for the testset. In the graph, the points represent the MR slices, and the black line is the regression line. The R-squared is 0.984 and the slope of the line is 0.992.
Figure 4 shows 3D tumor rendering of untreated and treated groups. The change of shape and size of tumors can be monitored through the time points.
Disscusion and Conclusion
The proposed segmentation application represented accurate tumor segmentation on untreated and treated groups. These results indicated that the proposed application could monitor tumor growth and the therapeutic effect of Doxorubicin. Also, tumor volume above 2mm3 can be segmented by the application, and the output, reconstructed to 3D tumor rendering can guide tumor characteristics. As a result, the proposed segmentation application can be able to apply to orthotopic mouse model research.Acknowledgements
This work was partially supported by grants from the National Research Foundation of Korea of the Korean government (Nos. 2018M3C7A1056887 and 2022R1A2C2011191). This research was supported by the 2021 Joint Research Project of the Institute of Science and Technology and was also supported by a grant from the Korea Healthcare Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant No: HI14C1135).References
1. Pillar, N., Polsky, A.L., Weissglas-Volkov, D. et al. Comparison of breast cancer metastasis models reveals a possible mechanism of tumor aggressiveness. Cell Death Dis 9, 1040 (2018). https://doi.org/10.1038/s41419-018-1094-8
2. Dutta, K.; Roy, S.; Whitehead, T.D.; Luo, J.; Jha, A.K.; Li, S.; Quirk, J.D.; Shoghi, K.I. Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary. Cancers 2021, 13, 3795. https://doi.org/10.3390/cancers13153795
3. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
4. He, Kaiming, et al. "Deep residual learning for image recognition." Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.APA
Figures
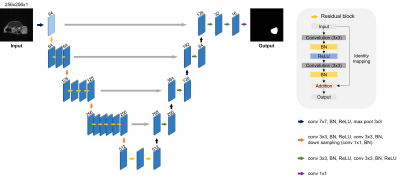
Figure 1. U-Net with ResNet34