4356
Brain tumor segmentation using out-of-distribution feature from normal MR images
Namho Jeong1, Beomgu Kang1, and Hyunwook Park1
1Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of
1Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Tumor
Abundant data of healthy subjects is available and thus it would be helpful if normal data can boost the brain tumor segmentation. We proposed the out-of-distribution (OOD) feature based on the learned distribution of normal data to discriminate abnormal tumors. Once the neural network was trained to synthesize other contrast MR images with normal data, it failed to properly synthesize the tumor tissues. This OOD characteristic was used for the generation of new feature that can help the tumor segmentation. It was verified that the OOD features contributed to an improvement of segmentation through experiments.Introduction
Automatic medical image segmentation is important since it can significantly reduce the cost of manual annotation in clinical practice. Recent advances in deep learning have shown the powerful ability of segmentation using multi-contrast MR images. However, most supervised learning methods focused on abnormal MR images to improve their performance, although normal MR images are more abundantly available. Therefore, it would be beneficiary if normal data can boost brain tumor segmentation. We developed the out-of-distribution (OOD) feature focusing on the normal multi-contrast MR images that can discriminate the abnormal tissues. A two-step framework was designed for class-specific tumor segmentation from multi-contrast MR images using the OOD features.Method
We used the multimodal BraTS challenge dataset, consisting of four MRI contrasts such as T1-weighted (T1w), T1 contrast-enhanced (T1-Gd), T2-weighted (T2w), and T2 fluid-attenuated inversion recovery (T2-FLAIR) volumes for this experiment2. In this study, we propose a two-step framework for the semantic segmentation of brain tumor tissues. In the first step, neural networks for synthesis are trained only with MR images of healthy people, and then MR images of the patients are given as input to obtain the OOD features. In the second step, semantic segmentation is performed with the OOD features obtained in the previous step as additional input. The class-specific segmentation, not multiclass segmentation, was designed to give different attention values when segmenting enhancing tumor (ET), necrosis/non-enhancing tumor (NCR/NET), and whole tumor (WT) independently. The deep neural networks of the two steps were modified from U-Net3.For the first step, deep neural networks are designed to synthesize each contrast of BraTS images from the other three contrast images, and trained with only the normal data as shown in the training phase as follows (Fig. 2):
$$\hat{x}_i = f_i(\tilde{X}_i), i=0,1,2,3$$
where $$$\hat{x}_i$$$ represents a synthesized MR image and $$$f_i$$$ represents a neural network for synthesizing $$$\hat{x}_i$$$. $$$\tilde{X}_i$$$ is a set of three images excluding $$$x_i$$$ out of $$$\{ x_0,x_1,x_2,x_3 \}$$$. $$$x_0,x_1,x_2,$$$ and $$$x_3$$$ are respectively T1w, T1-Gd, T2w, T2-FLAIR images in this study. A loss function for the first step is applied as a summation of L1 loss and structural similarity (SSIM) loss between a synthesized normal image $$$\hat{x}_i$$$ and a given normal image $$$x_i$$$ as follows4:
$$Loss1_i = L(\hat{x}_i,x_i)=L_1(\hat{x}_i,x_i)+\lambda*SSIM(\hat{x}_i,x_i)$$
SSIM is a perceptual metric that compares the brightness, contrast, and structure of two images, so it is possible to obtain a synthesized image more similar to the original image with additive SSIM loss and $$$\lambda$$$ of 0.1. At the test phase, if an input image including tumor tissues is given to the trained network, the region consisting of tumor tissues may not be properly synthesized. Meanwhile, the normal brain regions of target contrast can be synthesized well from three other contrast images, since the networks were trained with normal brain images.
In the second step, semantic segmentation of tumor tissue is performed using the results from the previous step. The extracted features are concatenated with the input multi-contrast images and utilized for the class-specific segmentation as shown in Fig. 1. The class-specific segmentation is performed for each class of tumor, and attention modules are added to all three pathways. The intensity difference of tumor core including enhancing tumor and necrosis/non-enhancing tumor is prominent in the T1-contrast enhanced images as shown in Fig. 3. Similarly, pattern differences in whole tumor are noticeable in T2-weighted or T2-FLAIR images. It is intended to put greater weight on useful information and lighter weight to suppress unnecessary information through convolutional block attention modules (CBAM). A loss function for the second step is a summation of dice loss and cross-entropy loss between a predicted segmentation map $$$\hat{y}_j$$$ and its label $$$y_j$$$ as follows:
$$Loss2_j = Dice(\hat{y}_j, y_j)+BCE(\hat{y}_j,y_j)$$
where the dice loss is a negative dice similarity coefficient and BCE is an abbreviation of the binary cross-entropy loss. $$$y_0,y_1,$$$ and $$$y_2$$$ are a ground truth segmentation map for ET, NCR/NET, and WT, respectively.
Experiments and Results
We evaluated our proposed method using the dice coefficient comparing with nnU-Net which took 1st place in the BraTS challenge 20205. The test dataset consisted of 57 patients, of which 41 cases were high-grade glioma (HGG) patients with relatively severe tumors and the other 16 cases were low-grade glioma (LGG) patients. As shown in Fig. 4, the overall performance of our proposed method shows improvements on the target regions over nnU-Net and proves that the OOD features obtained from the 1st step contributed to the improvements. Also, it is confirmed that the obtained OOD features of T1-Gd images show particularly high intensity in enhancing tumor area as shown in the 2nd column of Fig. 3, which could serve as a key feature of the attention modules.Conclusion
In the proposed method, we utilized the prominent features of tumor voxels in MR images such as the hyper-intensity of enhancing tumor tissues by the Gadolinium-based contrast agents. Our work described the two-step framework for semantic segmentation, which could provide the OOD features similar to the ground truth label. It is clear that the performance of segmentation was improved by both the OOD features and the CBAMs.Acknowledgements
This research was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT; the Ministry of Trade, Industry and Energy; the Ministry of Health and Welfare; the Ministry of Food and Drug Safety) (project number: 1711138003, KMDF-RnD KMDF_PR_20200901_0041-2021-02).References
1. Hendrycks, D., & Gimpel, K. (2016). A baseline for detecting misclassified and out-of-distribution examples in neural networks. arXiv preprint arXiv:1610.02136.2. Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., ... & Lanczi, L. (2014). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging, 34(10), 1993-2024.
3. Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
4. Zhao, H., Gallo, O., Frosio, I., & Kautz, J. (2016). Loss functions for image restoration with neural networks. IEEE Transactions on computational imaging, 3(1), 47-57.
5. Isensee, F., Jäger, P. F., Full, P. M., Vollmuth, P., & Maier-Hein, K. H. (2020, October). nnU-net for brain tumor segmentation. In International MICCAI Brainlesion Workshop (pp. 118-132). Springer, Cham.
6. Kim, B., Kwon, K., Oh, C., & Park, H. (2021). Unsupervised anomaly detection in MR images using multicontrast information. Medical Physics, 48(11), 7346-7359.
Figures
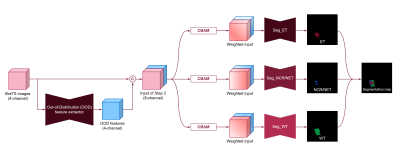
An overview of the proposed
two-step framework for brain tumor segmentation.
(Seg: segmentation, CBAM: Convolutional block
attention module)
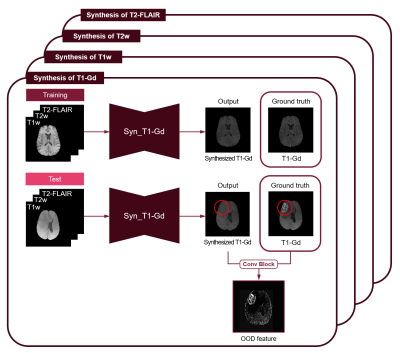
An overview
of OOD feature extractors trained only with normal MR images, showing a case for
T1-Gd images as a representative.
(Syn: synthesis)
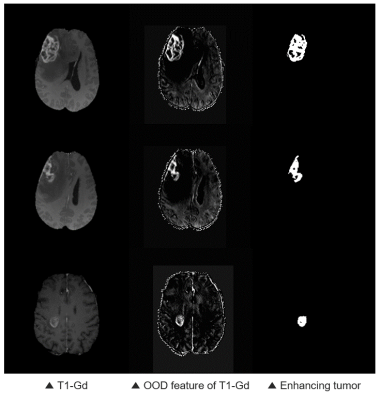
Examples of an OOD feature of a
T1-Gd image.
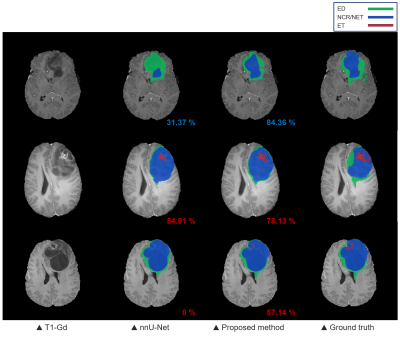
Qualitative results of brain
tumor segmentation from the nnU-Net and the proposed method. The numbers on the
bottom right of each segmentation map are the dice similarity coefficients of
ET (red) and NCR/NET (blue).
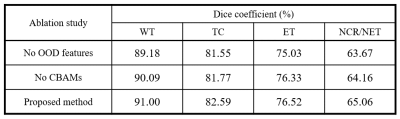
An ablation study of the proposed
segmentation method.
DOI: https://doi.org/10.58530/2023/4356