4353
A novel federated learning framework for accurate and secure multi-center MS lesion segmentation1the School of Computer Science, University of Sydney, Sydney, Australia, 2Brain and Mind Centre, University of Sydney, Sydney, Australia, 3Sydney Neuroimaging Analysis Centre, Sydney, Australia, 4the School of Biomedical Engineering, University of Sydney, Sydney, Australia, 5Sydney Imaging, University of Sydney, Sydney, Australia, 6the School of Electrical and Information Engineering, University of Sydney, Sydney, Australia
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation
Multiple sclerosis (MS) is a neurodegenerative disease of the central nerve system (CNS), which has the potential to cause a neurological disability, particularly for young adults. Recently, deep learning-based techniques are important for MS diagnosis and treatment, since they can segment the lesions caused by MS automatically and accurately. However, their applicability to multi-center scenarios is limited, due to the privacy and security issues in data sharing. To tackle these limitations, a decentralized deep learning framework is designed in this work, which can bring accurate multi-center MS lesion segmentation performance without sharing the raw data.Introduction
Multiple Sclerosis (MS) is a neurodegenerative disease of the central nervous system, featured by focal lesions. Accurate segmentation of MS lesions in magnetic resonance imaging (MRI) is critical in monitoring disease progression and evaluating treatment efficacy. With the recent advances in artificial intelligence (AI) techniques, deep learning methods have been widely employed for MS lesion segmentation achieving outstanding performance1. However, their applications in multi-center scenarios remain limited by sample size and often require multicenter collaborations. inevitably, the sharing of raw data and annotations across centers, giving raise to data security and privacy concerns. On the other hand, multi-center studies are crucial for accurate segmentation thanks to the exploitation of the inherent data variability from different centers2. To address the aforementioned limitations, we design a novel federated learning framework particularly for multi-center MS lesion segmentation with an accurate performance while maintaining data privacy.Material and Method
A multi-center MS lesion segmentation dataset from three different healthcare centers in Sydney with real-world clinical acquisitions is collected for this study. Specifically, 54 MRI cases are acquired using a GE Discovery 3T scanner (C1); 21 MRI cases were obtained using a Philips Ingenia 3T scanner (C2), and 30 cases are acquired using a Siemens Skyra 3T scanner (C3). The pre-processing steps included skull striping and N4 bias correction, and all the lesion masks were labeled by two expert neuroimage analysts at the Sydney Neuroimaging Analysis Centre. Only FLAIR images were employed in the experiments.The overview of the proposed Federated MS lesion Segmentation (FedMSSeg) method is shown in Figure 1. Our method extends the FedAVG method3. First, each center trains an AI model for MS lesion segmentation using their local data and in a fully-supervised manner. Specifically, a 3D U-Net4, the current state-of-the-art deep-learning model for medical image segmentation, is employed as the main segmentation model. Second, each center sends its model to a central server after local training. Third, the central server averages the weights of all the local models to generate an aggregated model. Finally, the aggregated model is sent back to each local center for the next round of federated learning. During inference, the segmentation model in each center is directly validated on their corresponding testing data.
To further improve this traditional federated learning method on MS lesion segmentation, two strategies are further designed. First, the parameters in the batch normalization layers of the AI models are not shared with central servers for aggregations. Second, an entropy-based aggregation strategy is designed in the central server to aggregate the model weights based on their segmentation uncertainty. For models with higher uncertainty, a low weight is assigned during aggregation.
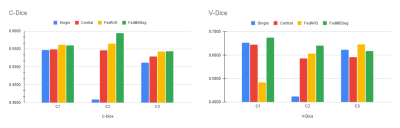
The models are trained and tested in a 3-fold cross-validation manner. To evaluate the model’s segmentation performance, the case-level and voxel-level Dice scores are reported. Case-level Dice (C-Dice) score is the average Dice score subject-wise, and the voxel-level Dice (V-Dice) score is computed over all the aggregated voxels from all the lesion predictions.
Results
We report the results for different multi-center MS lesion segmentation strategies:1) Single-center training. Each center trains and validate its models locally, without any communication procedures with other centers.
2) Centralized training. All centers share their data to train a model.
3) FedAVG. The traditional federated learning strategy.
4) FedMSSeg. Our proposed federated learning framework.
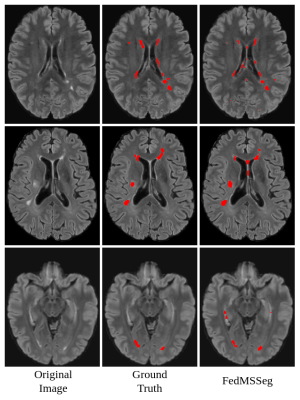
We obtain an overall C-Dice and V-Dice of our FedMSSeg of 0.57 and 0.64, respectively, outperforming the other three strategies: single-client training (0.49 / 0.57), centralized training (0.54 / 0.61), and FedAVG (0.56 / 0.58). The performance per center is shown in Figure 2. The results indicate our proposed federated learning strategy can outperform other comparison methods by a large margin, without sharing the raw data. Figure 3 shows the qualitative examples of our FedMSSeg.
Discussions
Our proposed FedMSSeg has outperformed the traditional federated learning method FedAVG. First, according to the previous studies5, the parameters within the batch normalization layers of each local model inherit the domain-specific knowledge of the training data in this specific center. Directly aggregating the multi-center domain-specific information in FedAVG brings feature shift to the local models and further limits their performance. Our model-sharing strategy without considering the domain-specific information will further enhance the models’ generalization ability. Second, due to the distinct characteristic of the multi-center MS lesion segmentation data, the local models collected by the central server have different segmentation abilities. Our entropy-based aggregation strategy has been designed to downgrade the attribute of the local models with lower segmentation confidence to maintain the aggregated model’s overall accuracy. By further outperforming the centralized training, our method is indicated to be effective while maintaining data privacy.Conclusion
We propose a novel decentralized deep learning framework for accurate multi-center MS lesion segmentation without sharing the raw data. Given the state-of-the-art performance on the datasets from real-world routine clinical data on lesion segmentation, future work will focus on other diseases to explore the utility and performance on other neuroimaging applications.Acknowledgements
The authors acknowledge the funding support by the Australia Medical Research Future Fund under Grant (MRFFAI000085), and the assistance from the Sydney Neuroimaging Analysis Centre.References
[1] Y. Ma et al., "Multiple Sclerosis Lesion Analysis in Brain Magnetic Resonance Images: Techniques and Clinical Applications," in IEEE Journal of Biomedical and Health Informatics, vol. 26, no. 6, pp. 2680-2692, June 2022, doi: 10.1109/JBHI.2022.3151741.
[2] S. Aslani et al., "Scanner Invariant Multiple Sclerosis Lesion Segmentation from MRI," 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020, pp. 781-785, doi: 10.1109/ISBI45749.2020.9098721.
[3] B. McMahan, et al. "Communication-efficient learning of deep networks from decentralized data." Artificial intelligence and statistics. PMLR, 2017.
[4] Ö Çiçek, et al. "3D U-Net: learning dense volumetric segmentation from sparse annotation." International conference on medical image computing and computer-assisted intervention (MICCAI). Springer, Cham, 2016.
[5] X. Li et al., "FedBN: Federated Learning on Non-IID Features via Local Batch Normalization," International Conference on Learning Representations (ICLR), 2021