4352
A Non-linear, Tissue-specific 3D Data Generative Framework for Deep Learning Segmentation Performance Enhancement
Soumya Ghose1, Chitresh Bhushan1, Dattesh Shanbhag2, Desmond Teck Beng Yeo3, and Thomas K. Foo3
1AI and Computer Vision, GE Research, Niskayuna, NY, United States, 2GE Healthcare, Bangalore, India, 3Biology & Physics, GE Research, Niskayuna, NY, United States
1AI and Computer Vision, GE Research, Niskayuna, NY, United States, 2GE Healthcare, Bangalore, India, 3Biology & Physics, GE Research, Niskayuna, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation, Vertebra segmentation, T2w, Deep Learning, Intensity Transformation
Deep learning (DL) models have been successful in solving segmentation problems that involve large, balanced, and labeled datasets. However, in the medical imaging domain, it is rare to find manually annotated datasets that capture the entire spectrum of heterogeneity. Novel datasets with significantly different intensities than training datasets, may adversely affect DL model performance. In this work, we present a hybrid framework for tissue-specific, non-linear intensity transformation of pediatric T2w images similar to that of the adults training dataset and demonstrate an improved performance for vertebra segmentation of pediatric datasets without the need for DL network re-training/re-tuning.INTRODUCTION
Deep learning (DL) models have proven successful in solving classification and regression problems that involve sufficiently large, balanced and labeled datasets1. In the medical imaging domain however, it is rare to find large, balanced and manually labelled datasets that capture the entire spectrum of heterogeneity inherent in the observations of interest. In the presence of novel datasets that significantly differ from training datasets in terms of appearance, contrast, shape (organs/structures of interest) and field of view, model performance may be adversely affected. Current DL schemes assume that all samples are available during the training phase. Therefore, after a DL network is trained with a primary training dataset and deployed for use, it may be necessary to re-train the network parameters with a more diverse training dataset if the distribution of the novel datasets observed in the field significantly deviates from that of the primary training dataset. Such re-training can be expensive as it involves large amount of new data acquisition and manual annotations to create ground truth labels. To mitigate the challenges associated with novel datasets, in this work, we present a hybrid framework of un-supervised and supervised learning to perform non-linear transformation of intensity such that the novel images have similar intensity profiles to that observed in the primary training dataset. We demonstrate that the performance of vertebra segmentation on the novel (pediatric) datasets did not degrade, which allows the existing (adults) DL model to perform without failures and without re-training with novel datasets. Compared to generative adversarial networks (GANs) for intensity transformation/generation2 that often requires large, paired dataset for learning the transformation parameters, our hybrid model learns from a single dataset without any manual intervention/annotation and GPU requirement resulting in minimum disruption of the workflow.METHOD
Clinical T2w MR images were chosen to build our hybrid non-liner intensity transformation framework. One T2w MR image from the adult training dataset and one T2w MR image of the new pediatric dataset were chosen for learning the intensity transformation/prediction. Learned transformation was applied to new images for validation. The hybrid approach can be broadly divided into two parts: (a) an unsupervised probabilistic modeling of the tissue types from T2w images followed by (b) non-linear regression-based transformation of T2w intensities to match intensity similar to training datasets. For training the model, one T2w image from the training dataset and one T2w image from the new dataset with significant intensity variations were used. For training the model, an expectation-maximization (EM)3 based clustering was performed on the each of the T2w MRIs to automatically create four clusters from intensities and identify the dense bone/air, vertebra/muscle and fat tissue classes in each of the T2w MRIs. Non-linear random forest regression models4 for individual tissue types/classes were used to transform the intensities similar to training images. Probabilities of individual tissue classes were used as features from the new dataset and adult T2w intensity as ground truth to predict tissue intensity similar to the training dataset. Three separate random forest regression models each corresponding to vertebra/muscle, fat and bone/air of 100 trees of depth three were created during training. During validation, given a new patient MRI image from the test/new dataset, an EM based clustering identified the vertebra/muscle, fat and air/bone. Tissue-specific regression models of the vertebra/muscle, bone and the fat generated in the training stage were used to transform T2w MR intensities of the test case to match the training intensities (Fig. 1).RESULTS
In Fig. 2, we show examples of vertebra segmentation of pediatric cases in which tissue intensities significantly differ from the adult cases. A 3D UNet segmentation model trained with adult images fails to accurately segment most vertebras due to intensity differences. The same segmentation model performance improves significantly after non-linear transformation of pediatric T2w contrast similar to adult T2w MRI using our tissue-specific, non-linear intensity transformation model.DISCUSSIONS & CONCLUSIONS
This is a preliminary study that shows non-linear, tissue-specific intensity transformation between pediatric and adult vertebra images improve DL segmentation model performance. No manual annotation or intervention, DL re-training/re-tuning were required for the process. Tissue transformation model was learnt from two T2w MRIs which significantly reduces data burden of learning such transformation models as typically required in GANs. We believe such tissue-specific, non-linear intensity transformation would improve performance of DL models and allow learning with less manually annotated data, significantly reducing data and time involved in building a robust DL models for real-world applications in medical imaging domain.Acknowledgements
No acknowledgement found.References
[1] Chai J, Zeng H, Li A, Ngai EWT. Deep learning in computer vision: A critical review of emerging techniques and application scenarios, Machine Learning with Applications, 2021; 6:100134.
[2] Aggarwal A, Mittal M, Battineni G. Generative adversarial network: An overview of theory and applications, International Journal of Information Management Data Insights. 2021; 1(1):100004.
[3] Moon TK. The expectation-maximization algorithm, in IEEE Signal Processing Magazine, 1996;13(6): 47-60.
[4] Breiman L. Random Forests. Machine Learning, 2001; 45: 5-32.
Figures
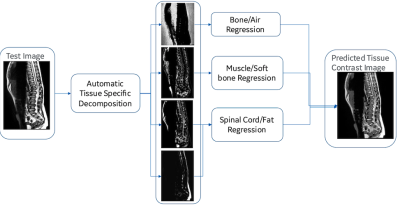
Figure 1 Tissue specific regression models are used for tissue contrast transformation T2w MRI of pediatric scan as pseudo adult scans.
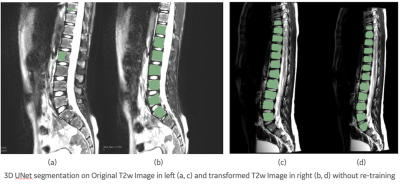
Figure 2 DL prediction on pediatric T2w image on the left, prediction on transformed T2w image on the right for the same slice. In (a) and (c) we can observe under segmentation getting better using the transformed T2w images in (b) and (d). We further observe better vertebra separation comparing (c) and (d). No re-training/re-tuning of the DL model was necessary in the process.
DOI: https://doi.org/10.58530/2023/4352