4349
Automated deep learning based segmentation of abdominal adipose tissue on whole-body MRI in a population-based study of adolescents1Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, Netherlands, 2Image Analysis, German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany, 3Department of Pediatrics, Erasmus MC, Rotterdam, Netherlands
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation, Dixon MRI
This study is embedded in the Generation R Study, a population-based prospective cohort study in the Netherlands. Whole-body Dixon MRI scans were performed at age 13 years. A previous neural network (CDFNet) has been published that was trained on adults. We aimed to retrain this network on our MRIs in the Generation R Study. The obtained segmentations are in strong agreement with expert-generated manual segmentations and can therefore greatly reduce the manual workload. Therefore, for accurate abdominal fat quantification, segmentation of both subcutaneous and visceral adipose tissue, on Dixon MRIs of adolescents is feasible with a retrained convolutional neural network.INTRODUCTION
Childhood obesity has become a major health problem worldwide 1. Several MRI techniques have been proposed to visualize body fat compartment, such as T1-weighted imaging by spin-echo or gradient-echo techniques or chemical shift selective imaging 2. Furthermore, Dixon MRI techniques, based on the difference between the resonance frequencies of protons bound in water and fat, were proposed to obtain both registered water and fat images, which have been shown to be useful for analysis of adipose tissue and muscles 3. Recently, fully convolutional neural network (F-CNN) has been described as state-of-art techniques for medical image segmentation 4. Promising results using CNNs to segment and quantify abdominal adipose tissue depots have been reported 5-8. Estrada et al. proposed FatSegNet 9, a fully automated pipeline to accurately segment adipose tissue inside a consistent anatomically defined abdominal region. However, the method has not been tested on a population cohort of adolescents. In this work, we aimed to evaluate an automated deep learning method for segmentation of subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) volumes on whole‑body MRI scans in adolescents.METHODS
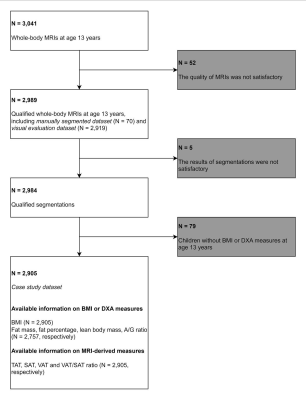
This study was conducted within Generation R Study, a population-based cohort study. A total of 3,041 whole-body Dixon MRIs from adolescents aged 13 years were used for abdominal adipose tissue segmentation (Figure 1). A previously presented Competitive Dense Fully Convolutional Network (CDFNet) was retrained on 62 individuals (training dataset) in a cross-validation approach. Segmentation accuracy was assessed via Dice score and volumetric similarity (VS) on eight individuals (testing dataset). The performance of the retrained CDFNets were compared against the model developed by Langner et al. 7, and the original model by Estrada et al. 9. A final model was implemented using all subjects from the training dataset.Furthermore, a subset of 504 automated segmentations was evaluated visually by two experienced observers scoring under- and over-segmentation rate on a scale 0-3, using intraclass correlation coefficient (ICC) to assess inter-observer agreement.
Finally, a case study was conducted to correlate MRI measurements with dual-energy X-ray absorption measurements and body mass index (BMI).
RESULTS
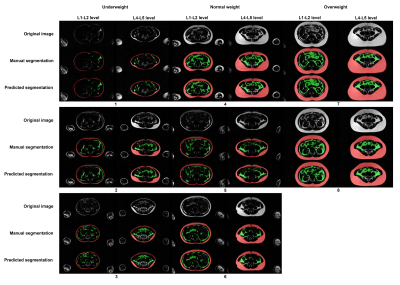
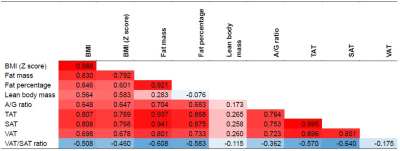
We observed that all retrained cross-validation models (CDFNet-S1 to S4) highly accurately segment SAT (Dice = 0.95, VS = 0.98). Moreover, the Estrada et al. (Dice = 0.92, VS = 0.96) and Langner et al. (Dice = 0.79, VS = 0.93) methods also perform well on SAT segmentation. For the more challenging task of VAT recognition, the retrained CDFNet models (Dice = 0.84, VS = 0.93) drastically outperform both Estrada et al. (Dice = 0.64, VS = 0.70) and Langner et al. (Dice = 0.66, VS = 0.92). Finally, the final model has comparable results when segmenting SAT (Dice = 0.95, VS = 0.98) and VAT (Dice = 0.85, VS = 0.93) as the cross-validation models (Figure 2, 3). The mean under- and over-segmentation rate of two observers and the agreement between them were 2.92 (ICC 0.931) and 2.99 (ICC 0.922) for SAT, and 2.99 (ICC 0.956) and 2.95 (ICC 0.778) for VAT respectively, indicating excellent segmentation quality across individuals (Figure 4). The correlation coefficients of VAT with SAT, fat mass and BMI were 0.851, 0.801 and 0.698, respectively (Figure 5).DISCUSSION
We retrained and evaluated a F-CNN (CDFNet) on MR scans acquired in a cohort of children. Through quantitative and qualitative evaluations, the retrained method showed a strong agreement with the ground truth for SAT and VAT. Our work demonstrated that the retrained CDFNet is effective in segmenting abdominal fat depots in whole-body Dixon MRIs of adolescents. The retrained method showed a strong agreement with the ground truth, and showed good generalizability to different BMI categories in the age group of 13 years. Furthermore, SAT segmentation showed higher performance than VAT segmentation based on the Dice score and VS on all evaluated models. Regarding computation time, it took approximately 15 seconds for one abdominal MRI scan to be fully processed using the retrained method on an NVIDIA 2080 Ti GPU with 11 GB of memory. This short time makes the method suitable for processing of large cohorts, and potentially for use in clinical routine settings. This study also facilitates future studies into abdominal fat distribution and its relationship with other obesity measures or clinical outcomes in adolescents.CONCLUSION
We retrained and evaluated the CDFNet model on whole-body MRIs of adolescents and accurately assessed abdominal SAT and VAT volumes on participants from Generation R.Acknowledgements
The Generation R study is managed by the Erasmus Medical Centre in close collaboration with the School of Law and the Faculty of Social Sciences at the Erasmus University, Rotterdam, the Municipal Health Service, Rotterdam area, and the Stichting Trombosedienst and Artsenlaboratorium Rijnmond (Star-MDC), Rotterdam. We gratefully acknowledge the contribution of children and their parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam.
References
1. Kumar S, Kelly AS. Review of childhood obesity: from epidemiology, etiology, and comorbidities to clinical assessment and treatment. Paper presented at: Mayo Clinic Proceedings2017.
2. Baum T, Cordes C, Dieckmeyer M, et al. MR-based assessment of body fat distribution and characteristics. European journal of radiology. 2016;85(8):1512-1518.
3. Ma J. Dixon techniques for water and fat imaging. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2008;28(3):543-558.
4. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Medical image analysis. 2017;42:60-88.
5. West J, Dahlqvist Leinhard O, Romu T, et al. Feasibility of MR-based body composition analysis in large scale population studies. PloS one. 2016;11(9):e0163332.
6. Newman D, Kelly‐Morland C, Leinhard OD, et al. Test–retest reliability of rapid whole body and compartmental fat volume quantification on a widebore 3T MR system in normal‐weight, overweight, and obese subjects. Journal of Magnetic Resonance Imaging. 2016;44(6):1464-1473.
7. Langner T, Hedström A, Mörwald K, et al. Fully convolutional networks for automated segmentation of abdominal adipose tissue depots in multicenter water–fat MRI. Magnetic resonance in medicine. 2019;81(4):2736-2745.
8. Küstner T, Hepp T, Fischer M, et al. Fully automated and standardized segmentation of adipose tissue compartments via deep learning in 3D whole-body MRI of epidemiologic cohort studies. Radiology: Artificial Intelligence. 2020;2(5):e200010.
9. Estrada S, Lu R, Conjeti S, et al. FatSegNet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. Magnetic resonance in medicine. 2020;83(4):1471-1483.
Figures
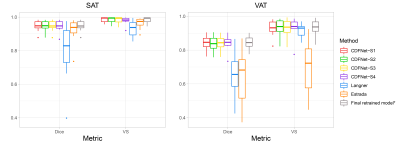
Dice score and Volumetric similarity for the four cross-validation splits and three methods. Results of SAT are shown on the left, and results of VAT are shown on the right. CDFNet-S1, CDFNet-S2, CDFNet-S3 and CDFNet-S4 represent four cross-validation splits. *Final retrained model was the best model out of 3 training experiments on the testing dataset, therefore it cannot be compared to the other methods that did not have access to the testing dataset. SAT: subcutaneous adipose tissue; VAT: visceral adipose tissue; VS: volumetric similarity.
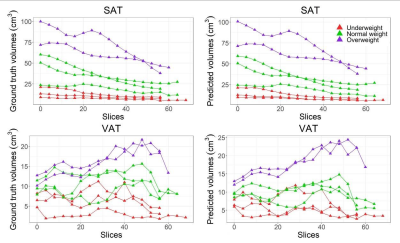
Comparison between the SAT and VAT volumes based on manual and automated segmentation in the testing dataset. The two figures on the left show the ground truth volumes of SAT and VAT, and the two figures on the right show the predicted volumes of SAT and VAT. From left to right in each figure means the direction from feet to head. SAT: subcutaneous adipose tissue; VAT: visceral adipose tissue.