4348
Cerebrovascular segmentation of mouse TOF-MRA using 3D U-Net
1AHU-IAI AI Joint Laboratory, Anhui University, Hefei, China, 2Institute of Artificial Intelligence, Hefei Comprehensive National Science Center, Hefei, China, 3State Key Laboratory of Brain and Cognitive Science, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China, 4University of Chinese Academy of Sciences, Beijing, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Vessels
Time-of-flight MR angiography (TOF-MRA) allows noninvasive visualization of the mouse intracranial vessels. However, there is no available tool for vessel segmentation in mouse 3D TOF-MRA. In this study, we developed an automated vessel segmentation method based on a convolutional neural network and multiscale vessel enhancement filtering algorithm. The method is potentially useful in the preclinical studies of cerebrovascular diseases.Introduction
Cerebrovascular disease models in mice are being widely used in preclinical studies to study phenotypes, causes, and treatments1. Time-of-flight MR angiography (TOF-MRA) allows noninvasive visualization of cerebral vessels. Accurate segmentation of vasculature is an essential step for researchers to analyze the pathogenic mechanisms and therapeutic efficacy of cerebrovascular diseases. However, the segmentation of mouse vasculature remains challenging due to the complex image context and vascular morphology. Recently, deep learning has achieved tremendous progress in the field of image segmentation. In this study, we proposed a method based on 3D U-Net and a multiscale vessel enhancement filtering algorithm3 to obtain automated vessel segmentation in 11.7T TOF-MRA of mice.Methods
The data from thirty mice were collected to train and evaluate the segmentation method. The MR images were acquired on an 11.7T animal scanner (Bruker, Biospec, Germany). The protocol of 3D TOF-MRA was: resolution = 0.10x0.10x0.10mm3, matrix = 256x256x180, TR = 12ms, TE = 2.21ms, FA = 20°.Before segmenting the vascular structure, the images in the dataset were preprocessed by RATS2 to extract brain regions (https://iibi.uiowa.edu/rats-rodent-brain-mri) and N4ITK4 bias correction to remove field inhomogeneity (http://hdl.handle.net/10380/3053). The vessels were enhanced by the multiscale filtering algorithm, with the parameters of sigma = 0.10 - 0.25 and number of steps = 8.
The patches with dimensions of 64x64x64 voxels were generated from the enhanced images. Interactive vessel segmentation was performed to acquire the labeled data for vessel segmentation. First, the image was filtered by using histogram-based thresholding. The threshold was chosen between 30% and 99% image intensity. Then, the vascular mask was manually adjusted to obtain the final labels. The basic structure of 3D-UNet for mouse cerebrovascular segmentation is represented in Figure 1. In the following task, 80 patches were used in the training stage, while 20 patches were used in the testing stage.
Result
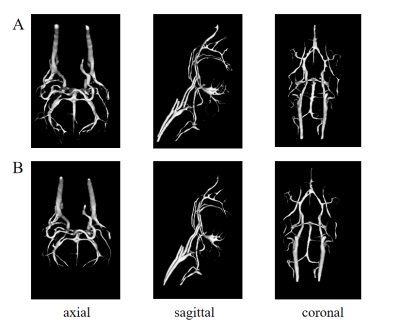
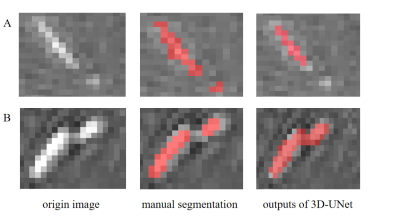
The surface rendering of the 3D vascular models is represented in Figure 2. The Dice similarity coefficient is 75.32%, with a false positive rate of 0.002, and a false negative rate of 0.161. For small vessels, detailed comparisons are shown in Figure 3. In most cases, manual segmentation is more accurate than 3D-UNet. However, the vessel labels missed in manual segmentation are sometimes corrected by 3D-UNet.Discussion
In this work, we proposed an automated vessel segmentation method to model mouse cerebral vasculature in TOF-MRA images, which used 3D-UNet and a multiscale vessel enhancement filtering algorithm. In Figure 2, the output of 3D-UNet achieves comparable results to manual segmentation in main arteries. The segmentation faults are mainly attributed to the false negatives in small vessels by 3D-UNet.In our experience, it is usually difficult and time-consuming for manual segmentation of TOF-MRA, especially when labeling small vessels. In Figure 3, the mismatches between U-Net outputs and manual labeling come from not only the false negative segmentation of U-Net but also the labeling faults missed by manual segmentation. These factors contribute to the relatively low dice coefficients. The segmentation algorithm still needs further improvements in dealing with small vessels. Nonetheless, considering the unavailability of an automated vessel segmentation tool in mouse 3D TOF-MRA images, our method has great value in the preclinical studies of neurovascular diseases.Acknowledgements
This work was supported by the National Natural Science Foundation of China (82271985, 82001804, 31730039), the National Science and Technology Innovation 2030 Major Program (2022ZD0211900, 2022ZD0211901), and the National Key Research and Development of China (2017YFC1307904).References
1. E. Sander Jr. Connolly, Christopher J. Winfree, David M. Stern, Robert A. Solomon, and David J. Pinsky. Procedural and strain-related variables significantly affect outcome in a murine model of focal cerebral ischemia. Neurosurgery, 38(3):–, 1996.
2. Ipek Oguz, Honghai Zhang, Ashley Rumple, Milan Sonka,RATS: Rapid Automatic Tissue Segmentation in rodent brain MRI,Journal of Neuroscience Methods,Volume 221,2014,Pages 175-182,ISSN 0165-0270.
3. Frangi, Ro & Niessen, W.J. & Vincken, Koen & Viergever, Max. (2000). Multiscale Vessel Enhancement Filtering. Med. Image Comput. Comput. Assist. Interv.. 1496.
4. N. J. Tustison et al., "N4ITK: Improved N3 Bias Correction," in IEEE Transactions on Medical Imaging, vol. 29, no. 6, pp. 1310-1320, June 2010, doi: 10.1109/TMI.2010.2046908.
Figures