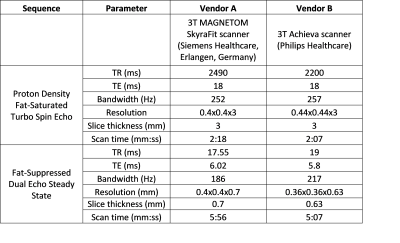4346
Novel Uunsupervised Segmentation of Bone Marrow Edema-Like Lesions using Bayesian Conditional Generative Adversarial Networks1Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States, 2Department of Biomedical Engineering, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, United States, 3Cleveland Clinic, Department of Computer and Data Sciences, Case Western Reserve University, Cleveland, OH, United States, 4Department of Electrical, Computer, and Systems Engineering, Case Western Reserve University, Cleveland, OH, United States, 5Department of Diagnostic Radiology, Imaging Institute, Cleveland Clinic, Cleveland, OH, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation
Quantitative assessment of the bone marrow edema-like lesions and their association with osteoarthritis requires a consistent and unbiased segmentation method, which is difficult to obtain in the presence of human annotators. This study proposes an unsupervised approach using Bayesian deep learning and conditional generative adversarial networks that detects and segments anomalies without human intervention. The full pipeline has a lesion-wide sensitivity of 0.86 on unseen scans. This approach is expected to be generalizable to other lesions and/or modalities.Introduction
Bone marrow edema-like lesions (BMEL) have been associated with disease progression and patient symptoms (such as pain) in osteoarthritis1,2. BMEL are also commonly seen after acute injuries such as ACL injury and meniscal tear and their persistence may be associated with post-traumatic osteoarthritis (PTOA) development3-5. Previous studies on BMEL were primarily limited by semi-quantitative analysis or grading while quantitative analysis of these lesions may increase the power of evaluating such associations and may have higher sensitivity to detect longitudinal changes. A few studies with quantitative analysis of BMEL used manual or semi-automatic methods for BMEL segmentation6,7, which is time consuming and prone to intra-/inter-reader variation. In this study, we propose a novel, unsupervised method for the automatic segmentation of BMEL using a Bayesian conditional generative adversarial network (cGAN)8.Methods
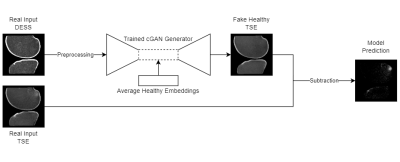
A total of 284 knee MRI scans were collected from two multi-site multi-vendor prospective cohorts: Multicenter Orthopaedics Outcomes Network (MOON)9 and Corticosteroid Meniscectomy Randomized Trial (CoMeT). Fat-suppressed dual echo steady state (DESS) and turbo spin echo (TSE) images with protocols listed in Table 1 were used for this study.Fig. 1 illustrates the overall model design. The cGAN model is first trained to translate healthy scans from the real input DESS to a fake output TSE. This is done by training the model only on ‘healthy’ scans without BMEL, so that it learns to generate a ‘healthy’ output regardless of the input10,11. The difference between the real unhealthy target TSE with BMELs and fake healthy TSE output is the predicted region(s) with BMEL. DESS and TSE are ideal sequences due to their difference in capturing BMEL signal, leading to a larger difference between the real and fake TSE images. These scans were preprocessed using rigid registration from DESS to TSE, cropped for femoral and tibial bone using an in-house, fully automatic software, and z-score normalization12.
The generator in the cGAN model uses a modified 2D U-Net13 with seven convolutional layers, three of which have random spatial dropout14, and the generator uses a modified PatchGAN15 classifier with four convolutional layers. In addition to the standard GAN losses, the L1 reconstruction loss were used as loss function for the generator.
4,467 DESS and TSE slice pairs without BMEL from 189 scans without metal artifacts were split 80-20 into training and validation sets. Afterwards, the features of each healthy scan are collected at the innermost layer of the generator, and their average (feature-wise arithmetic mean) replaces the corresponding features of each input scan during inference. This has a dampening effect on false positives, while preserving the spatial features of the input via the signature skip-connections in the U-Net architecture. Also, all dropout layers in the generator are enabled during inference, causing some random sets of the network weights to be ignored. Repeating the inference on the same input multiple times equates to a Monte Carlo simulation16 of the voxel-wise variance of the output. Regions with predicted lesions but also high variance are ignored since they are likely to be false positives17.
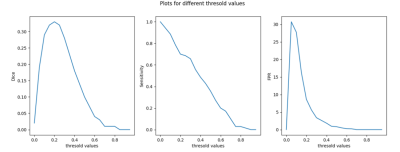
20 scans with BMEL were manually segmented in TSE images and used for testing, including the postprocessing pipeline for binary segmentation, which includes grayscale closing on the prediction and grayscale opening and selecting BMEL in central regions of the scans. To find the optimal threshold value, lesion-wise sensitivity and number of false positives per scan for 14 of the 20 scans were plotted against a range of threshold values applied to the difference between fake and real TSE images (Fig. 2). The remaining 6 were used for testing metrics.
Results
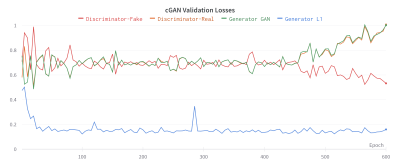
During training, the validation GAN losses were stable by epoch 400 before diverging, indicating overfitting (Fig. 3). The generator weights at epoch 400 were used for testing and evaluation.As shown in Fig. 4, the predictions and annotations agree on the presence and locations of BMELs, but do not agree on their exact boundaries. Unsurprisingly, the unsupervised model also discovers other lesions and high-intensity regions in the bone marrow that are not subchondral BMELs.
Based on Fig. 2, the threshold value of 0.2 was chosen. On the unseen 6 test subjects the unsupervised segmentation pipeline obtained a lesion-wise sensitivity and false positives per scan of 0.86, and 7.8 respectively.
Discussion
Due to the diffusive nature of BMELs, there are no ground-truth boundaries of the lesions, which constitutes a significant challenge for reliable and consistent BMEL segmentation. Previous studies reported significant differences in volume calculation using manual segmentation vs using computer assisted segmentation6,7. The lesion detection sensitivity of 0.86 was promising, but DICE score should be investigated in the future. After additional training with more data, the model can be used supplementarily in clinical practice with minimal cost. There are still many challenges that prevent it from being deployed in an end-to-end, fully unsupervised manner, but the results suggest the possibility of a more generalized solution that is applicable to a variety of anomalies and imaging modalities.Conclusion
This study illustrates the viability of unsupervised segmentation of BMELs using Bayesian cGANs and injected embeddings. With further improvement, reliable automated quantitative analysis of BMEL will greatly facilitate evaluation of this lesion in large clinical trials for osteoarthritis.Acknowledgements
The study was supported by NIH/NIAMS R01AR075422 and the Arthritis Foundation.References
1. Felson, David T., et al. "Bone marrow edema and its relation to progression of knee osteoarthritis." Annals of internal medicine 139.5_Part_1 (2003): 330-336.
2. Felson DT, Niu J, Guermazi A, et al. Correlation of the development of knee pain with enlarging bone marrow lesions on magnetic resonance imaging. Arthritis Rheum. 2007;56(9):2986-2992.
3. Papalia R, Torre G, Vasta S, Zampogna B, Pedersen DR, Denaro V, Amendola A. Bone bruises in anterior cruciate ligament injured knee and long-term outcomes. A review of the evidence. Open Access J Sports Med 2015;6:37-48.
4. Gong J, Pedoia V, Facchetti L, Link TM, Ma CB, Li X. Bone marrow edema-like lesions (BMELs) are associated with higher T1ρ and T2 values of cartilage in anterior cruciate ligament (ACL)-reconstructed knees: a longitudinal study. Quant Imaging Med Surg. 2016 Dec;6(6):661-670. doi: 10.21037/qims.2016.12.11. PMID: 28090444; PMCID: PMC5219965.
5. Filardo G, Kon E, Tentoni F, Andriolo L, Di Martino A, Busacca M, Di Matteo B, Marcacci M. Anterior cruciate ligament injury: post-traumatic bone marrow oedema correlates with long-term prognosis. Int Orthop 2016;40:183-90. 10.1007/s00264-015-2672-3
6. Preiswerk, Frank, et al. "Fast quantitative bone marrow lesion measurement on knee MRI for the assessment of osteoarthritis." Osteoarthritis and Cartilage Open 4.1 (2022): 100234.
7. Nielsen, Flemming K., et al. "Measurement of bone marrow lesions by MR imaging in knee osteoarthritis using quantitative segmentation methods–a reliability and sensitivity to change analysis." BMC Musculoskeletal Disorders 15.1 (2014): 1-11.
8. Mirza, Mehdi, and Simon Osindero. "Conditional generative adversarial nets." arXiv preprint arXiv:1411.1784 (2014).
9. Xie, D., et al. "Multi-vendor multi-site quantitative MRI analysis of cartilage degeneration 10 Years after anterior cruciate ligament reconstruction: MOON-MRI protocol and preliminary results." Osteoarthritis and Cartilage (2022).
10. Schlegl, T., Seebock, P., Waldstein, S. M., Schmidt-Erfurth, U., and Langs, G., “Unsupervised anomaly detection with generative adversarial networks to guide marker discovery,” in [International Conference on Information Processing in Medical Imaging], 146–157, Springer (2017).
11. Baur, C., Wiestler, B., Albarqouni, S., and Navab, N., “Deep autoencoding models for unsupervised anomaly segmentation in brain MR images,” in [International MICCAI Brainlesion Workshop], 161–169, Springer (2018).
12. Reinhold, Jacob C., et al. "Evaluating the impact of intensity normalization on MR image synthesis." Medical Imaging 2019: Image Processing. Vol. 10949. SPIE, 2019.
13. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
14. Tompson, Jonathan, et al. "Efficient object localization using convolutional networks." Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
15. Isola, Phillip, et al. "Image-to-image translation with conditional adversarial networks." Proceedings of the IEEE conference on computer vision and pattern recognition. 2017.
16. Gal, Y. and Ghahramani, Z., “Dropout as a bayesian approximation: Representing model uncertainty in deep learning,” in [International conference on machine learning], 1050–1059 (2016).
17. Reinhold, Jacob C., et al. "Finding novelty with uncertainty." Medical Imaging 2020: Image Processing. Vol. 11313. SPIE, 2020.
Figures