4345
ZTE segmentation of glenohumeral bone structure using deep learning
Michael Carl1, Kaustaub Lall2, Armin Jamshidi2, Eric Chang3,4, Sheronda Statum3,4, Anja Brau1, Christine B Chung3,4, Maggie Fung1, and Won C. Bae3,4
1General Electric Healthcare, Menlo Park, CA, United States, 2Electrical and Computer Engineering, University of California, San Diego, La Jolla, CA, United States, 3Radiology, University of California, San Diego, La Jolla, CA, United States, 4Radiology, VA San Diego Healthcare System, San Diego, CA, United States
1General Electric Healthcare, Menlo Park, CA, United States, 2Electrical and Computer Engineering, University of California, San Diego, La Jolla, CA, United States, 3Radiology, University of California, San Diego, La Jolla, CA, United States, 4Radiology, VA San Diego Healthcare System, San Diego, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation, ZTE, Deep learning
Evaluation of 3D bone morphology of the glenohumeral joint is necessary for pre-surgical planning. Zero echo time (ZTE) MRI provides excellent bone contrast, and we developed a deep learning model to perform automated segmentation of major bones (i.e., humerus and others) from ZTE to aid evaluation. Axial ZTE images of normal shoulders (n=31) acquired at 3T were annotated for training with a 2D U-Net, and the trained model was validated with testing data (n=10 normal shoulder, n=6 symptomatic). Testing accuracy was around 80 to 90% (Dice score) for either cohort, except for a few failed cases with very low scores.PURPOSE
Evaluation of patient-specific 3D morphology of the bone is crucial for pre-surgical planning.1, 2 Currently, CT imaging followed by 3D reconstruction and visualization is used. Glenohumeral joint (GHJ) osteoarthritis is one of the most common indicators for shoulder arthroplasty,3 and recent study on zero echo time (ZTE) MR imaging suggested the ability to depict bone contrast for the GHJ.4 Unfortunately, manual segmentation is currently needed to depict different joints, i.e., humeral head or glenoid. This study aims to develop an automated deep-learning based segmentation to separate these two structures to aid evaluation.METHODS
ZTE Shoulder Data: Training Data: We obtained ZTE shoulder dataset through an existing study where n=31 datasets of normal shoulders, acquired on General Electric 3-Telsa scanners, were available. The images were acquired in the axial plane using mixed scan parameters: TR=100 to 600 ms, TE=0.016 to 0.028 ms, field of view (FOV)=160 to 240 mm, matrix=256 to 512, slice thickness=0.7 to 1.2 mm, number of slices=90 to 200. Images were rescaled to 256x256 in-plane only, enhanced with either SCENIC or PURE intensity correction, and were inverted to show bone with high signal intensity. An example axial ZTE shoulder image is shown in Figure 1A. Testing Data: For testing, additional n=10 ZTE shoulder data from normal subjects and n=6 from symptomatic subjects (4 male 2 female; age 40 to 69 yo) with recent shoulder pain were obtained, with similar scan parameters as the training data.Annotation: All images were annotated using ImageJ. Raw images (Figure 1A) were loaded as a 3D stack, and annotated into three separate binary 3D images, representing (i) the background / air (Figure 1B), (ii) humeral head and humerus (Figure 1C), and (iii) all other tissues including soft tissue and bone (i.e., glenoid, acromion, etc., Figure 1D). Note that humerus segmentation was performed loosely around the structure, to avoid accidental cropping of bony structures.
Deep Learning Segmentation: We have implemented a 2D U-Net5 deep learning (DL) model (Figure 2) written in Matlab (R2021a) to perform segmentation of shoulder ZTE images. The model takes in a 256x256 image, uses 64 convolution filters with 3x3 kernel size, 2 convolution operations at each step, with an encoder depth of 5 (or 9 layers, 4 down-sampling followed by 4 up-sampling). The final output consists of 3 separate black and white images to be used as masks for the background (air), humeral head/humerus, and the remaining tissues. The model was trained to 120 epochs (~2 days) using the default setting (Adam optimizer, Shuffling image every epoch, batch size=8) on a Windows 10 PC with i7-10700K CPU, 32GB RAM, RTX3090 GPU with 24GB VRAM. Training results were good, reaching ~99% accuracy (Figure 3).
Accuracy: Compare vs. Manual Segmentation: Aforementioned ZTE Test Data on n=10 normal and n=6 symptomatic subjects were processed with the trained U-Net model to perform DL segmentation (inference takes ~2 min), and compared against manual segmentation. Dice score6 was calculated for segmentation of the humerus and the remaining tissues, separately. Compare vs. CT: We had an opportunistic dataset where both CT and ZTE MR data of a shoulder were available. The CT and ZTE data were first registered, and the registered ZTE data was DL segmented, while the CT data manually segmented to isolate the humerus. The segmented CT and ZTE data were overlaid to visualize the overlap.
RESULTS
When the testing data (Figure 3A) was DL segmented (Figure 3BC) and compared against manual segmentation (Figure 3DE), it could be seen that DL processing yielded satisfactory results, successfully segmenting the humeral head in most cases. Segmented images facilitate 3D visualization of the humeral head (Figure 3F) and glenoid (Figure 3G) for evaluation.Figure 4 shows Dice scores. Vast majority had high Dice scores of 80 to 90%, although there were a few that had markedly lower values (highlighted in pink). Testing data on normal subjects (Figure 4A) had Dice scores of 77% +/- 30% (mean +/- standard deviation) on humerus and 91% +/-10% on the remaining tissues. On the symptomatic subjects (Figure 4B), the number were similar, with Dice scores of 77% +/- 14% on humerus and 90% +/-6% on the remaining tissues. The DL segmentations worked satisfactorily for the majority of the cases, and the accuracy between different cohorts of subjects was similar.
When comparing ZTE (Figure 5A) and CT (Figure 5B) data from the same subject, the Dice score of humerus segmentation was 97%. We created a fused 3D rendering (Figure 5C), which shows an excellent overlap (white) between ZTE (purple) and CT (green).
CONCLUSION
We have successfully implemented DL model to segment the humerus and other tissues from ZTE shoulder images with varying scan parameters. This work may facilitate clinical evaluation of shoulder bones by eliminating manual post-processing. The present model is currently limited to axial images and there are failed cases without a clear cause, necessitating additional investigation.Acknowledgements
No acknowledgement found.References
- Bizzotto N, Tami I, Santucci A, Adani R, Poggi P, Romani D, Carpeggiani G, Ferraro F, Festa S, Magnan B. 3D Printed replica of articular fractures for surgical planning and patient consent: a two years multi-centric experience. 3D Print Med 2: 2, 2015. PMCID:PMC6036663
- Jehan S, Akhi Baig NM, Tavitian J. Treatment of Shoulder Dislocation with Greater Tuberosity and Glenoid Fractures. J Coll Physicians Surg Pak 26: 997-999, 2016.
- Macias-Hernandez SI, Morones-Alba JD, Miranda-Duarte A, Coronado-Zarco R, Soria-Bastida MLA, Nava-Bringas T, Cruz-Medina E, Olascoaga-Gomez A, Tallabs-Almazan LV, Palencia C. Glenohumeral osteoarthritis: overview, therapy, and rehabilitation. Disabil Rehabil 39: 1674-1682, 2017.
- Breighner RE, Endo Y, Konin GP, Gulotta LV, Koff MF, Potter HG. Technical Developments: Zero Echo Time Imaging of the Shoulder: Enhanced Osseous Detail by Using MR Imaging. Radiology 286: 960-966, 2018.
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 234-241, Year.
- Dice L. Measure of the Amount of Ecologic Association Between Species. Ecology 26: 297-302, 1945.
Figures
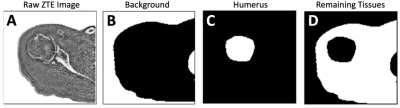
Figure 1. (A) Inverted ZTE shoulder images acquired in the axial plane were manually annotated (segmented) into (B) background, (C) humeral head / humerus, and (D) remaining tissues.
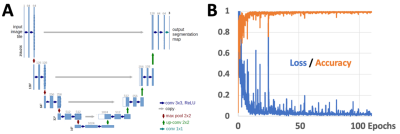
Figure 2. (A) Structure of our U-Net used in this study. (B) Training results of loss and accuracy.
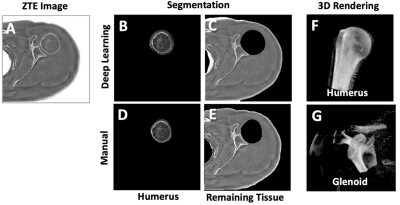
Figure 3. (A) ZTE test image was DL segmented to obtain images of humerus-only (B) and remaining tissue (C). These were compared against manually segmented humerus (D) and the remaining tissue (E). Segmented ZTE images facilitate 3D rendering and evaluation of the humeral head (F) and the glenoid (G).
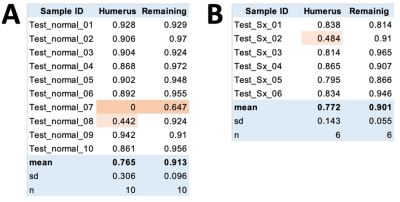
Figure 4. Dice scores between DL vs. manual segmentation on (A) normal and (B) symptomatic shoulders.
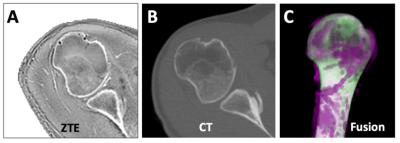
Figure 5. ZTE (A) and CT (B) data of the same subject was registered and segmented (using DL for ZTE, manually for CT). Segmented humerus was fused (C), showing overlapping regions as white, nonoverlapping regions in magenta for ZTE and green for CT.
DOI: https://doi.org/10.58530/2023/4345