4344
Spatial-Adaptive Deep Learning Model and Magnetic Resonance Fingerprinting for Segmentation and Quantitative Evaluation of Cervical Cancer1Radiotherapy and Imaging, The Institute of Cancer Research, London, United Kingdom, 2Radiology, The Royal Marsden Hospital, London, United Kingdom, 3Medical Imaging and Intervention, Chang Gung Memorial Hospital at Linkou and Chang Gung University, Taoyuan, Taiwan, 4Gynaecological Unit, The Royal Marsden Hospital, London, United Kingdom
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Cancer
Quantitative magnetic resonance imaging (qMRI) can provide additional information for diagnosis and response assessment, but adoption of multi-parametric qMRI techniques has been hindered by long acquisition times and labor-intensive processing steps. Magnetic resonance fingerprinting (MRF) provides quantitative maps in a single acquisition but MRF deployment in clinical studies still requires manual delineation of volumes of interest. A spatial-adaptive deep learning framework was developed to segment cervical cancer on MRI and quantify T1 relaxation times of the tumor pre- and post-radiotherapy treatment. Our results suggest that automated segmentation models may be promising tools for quantitative tumor evaluation and treatment response assessment.Background
Quantitative magnetic resonance imaging (qMRI) denotes mapping of underlying quantitative tissue properties (e.g. T1, T2 relaxation times)1. Multi-parametric qMRI methods suffer from long acquisition times, which hinder integration into routine clinical care. Recently, the introduction of magnetic resonance fingerprinting (MRF), for simultaneous estimation of these tissue properties, has shifted the paradigm in quantitative imaging2. While MRF promises to bring qMRI to cancer tumor analysis, it relies on manual delineation of regions of interest that increases the clinician workload. In this study, we propose a novel spatial-adaptive deep learning (DL)-based framework to automatically delineate cervical tumors on T2-weighted and quantify changes in T1 relaxation times between MRI pre- and post-radiotherapy (RT) measurements. Moreover, this pipeline offers an approach to bridge the semantic knowledge between two-dimensional (2D) diagnostic and three-dimensional (3D) RT planning MRI scans for improved segmentation outcome.Purpose
Develop and evaluate a novel and end-to-end deep learning framework for automatic segmentation and quantitative analysis of cervical cancer tumors pre- and post-treatment using semantic knowledge transfer and MRF respectively.Methods
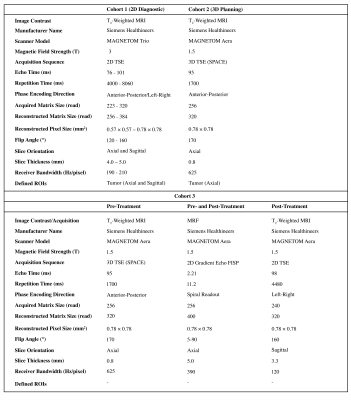
Patient Population and Imaging ProtocolsInstitutional review board approval was obtained for this study. Three cohorts were identified for tumor segmentation pre-training, transfer learning and evaluation. Cohort 1: 125 patients with cervical cancer undergoing 2D diagnostic abdomen-pelvis T2-weighted MRI (T2W) in axial and sagittal planes (MAGNETOM Trio, Siemens Healthcare, Erlangen, Germany). Cohort 2: 21 patients with 3D T2W MRI acquired for RT planning (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany). Cohort 3: 18 patients undergoing 3D T2W MRI for RT planning pre-treatment, from whom 13 patients also had 2D T2W MRI for post-treatment follow-up. The MRF measurements were acquired during both patient visits (9 axial slices, positioned at the center of cervical tumors) using a prototype Fast Imaging with Steady State Precession (FISP) sequence3 with Gadgetron reconstruction4 that returns maps of T1 and T2 relaxation times, and proton density M0. In this cohort, 16 patients had a second baseline MRF acquisition for assessment of repeatability (Figure 1).
Framework Architecture and Training
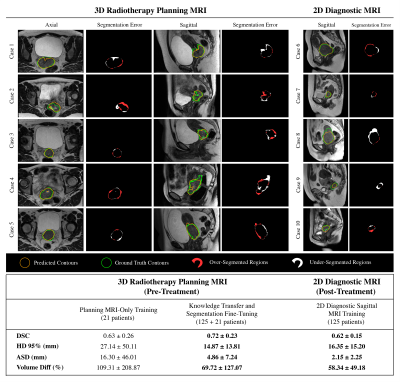
All images in cohort 1 were normalized (mean: 0, variance: 1) and resampled to an isotropic global space to match the resolution of images in cohort 2 (0.8×0.8×0.8 mm3). The axial and sagittal planes embed global semantic information of diagnostic images to transfer to planning MR images. A modified residual U-Net5 with 4 down- and up-sampling blocks and 2 residual units was developed to perform 3D patch-based (128×128×128) tumor segmentation (train: 75, validation: 25, test: 25 patients). The trained weights were used for segmentation fine-tuning of cohort 2 MR images (train: 13, validation: 4, test: 4 patients). The quantitative segmentation metrics were investigated after four-fold cross validation for target-only training and pre-training segmentation adaptation. The training for post-treatment tumor segmentation was implemented on sagittal MRI images of size (256×256×16) from cohort 1 to expand the field of view and reduce pre-processing workload. The segmentation volumes were generated by sliding window patch fusion and intensity averaging (75% overlap).
Magnetic Resonance Fingerprinting (MRF) T1 Measurements
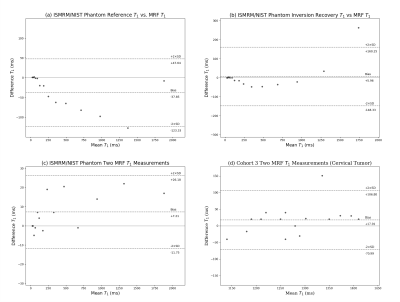
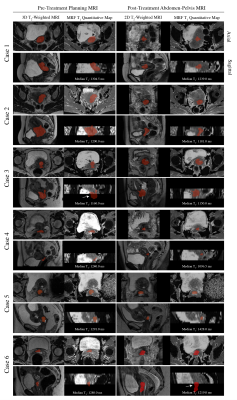
The MRF sequence was validated on the ISMRM/NIST MRI system phantom (CaliberMRI, Inc, Boulder, CO) by comparison with reference T1 values from the phantom manufacturer and comparison with T1 relaxation times estimated using inversion recovery turbo spin echo (IR-TSE) (10 inversion times) non-linear curve fitting6. For these comparisons, the median T1 values were extracted from the regions of interest (ROIs) delineated for each sphere. The T1 measurements of cervical tumors in cohort 3 were based on 16 patients who had two measurements during their first visit. The tumor contours were generated by the DL framework, and manually revised to account for internal tissue motions between scans. The schematic of the framework is shown in Figure 2.
Results
The segmentation results suggest that large-scale transfer learning from 2D diagnostic to 3D planning MRI improved the segmentation outcome compared with training without domain knowledge transfer (Figure 3). The MRF repeatability measurements for the ISMRM/NIST phantom and cohort 3 segmentations are shown in Figure 4. From 13 patients that underwent both pre- and post-MRF scanning, the cervical tumors were detected for 6 patients by the framework. From preliminary analysis, T1 relaxations in 5/6 patients were reduced after treatment. However, two-tail paired T-test statistics revealed no significant changes in these values (p>0.05) (Figure 5).Discussion and Conclusion
The rise of DL and rapid qMRI techniques offer the opportunity for automated quantitative measurements in clinical MRI. Quantitative properties from the MRF can be promising biomarkers in characterising RT treatment response in cancer7. We developed an end-to-end spatial-adaptive framework whereby cervical cancer can be delineated automatically and propagated to MRF scans for quantitative analysis. The key limitations are internal motion during the examination, leading to differences in tumor/organ position between the anatomical and qMRI data, and difficulty in segmenting some tumors. Another drawback is that the MRF acquisition consisted of nine 5-mm slices, due to constraints on acquisition time, and therefore did not cover the whole tumor in some cases. Future work will include qualitative radiologist assessments and automatic quantification of apparent diffusion coefficients (ADC) of cervical cancers. Additionally, multi-parametric MRI input for segmentation training will be investigated.Acknowledgements
This study represents independent research funded by the National Institute for Health and Care Research (NIHR) Biomedical Research Centre and the Clinical Research Facility in Imaging at The Royal Marsden NHS Foundation Trust and The Institute of Cancer Research, London, United Kingdom. The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. Gigin Lin received research funding from the Ministry of Science and Technology Taiwan (MOST 110-2628-B-182A-018). We are grateful to Vikas Gulani and Yun Jiang at the University of Michigan for supplying the MRF sequence.References
[1] Tofts P, editor. Quantitative MRI of the brain: measuring changes caused by disease. John Wiley & Sons; 2005.
[2] Ma D, Gulani V, Seiberlich N, Liu K, et al. Magnetic resonance fingerprinting. Nature. 2013; 495(7440):187-192.
[3] Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magnetic resonance in medicine. 2015;74(6):1621-31.
[4] Lo WC, Jiang Y, Franson D, et al. MR Fingerprinting using a Gadgetron-based reconstruction. InProc. ISMRM. 2018;3525.
[5] Kerfoot E, Clough J, Oksuz I, Lee J, King AP, Schnabel JA. Left-ventricle quantification using residual U-Net. In International Workshop on Statistical Atlases and Computational Models of the Heart. Springer, Cham 2018;371-380.
[6] Stupic KF, Ainslie M, Boss MA, et al. A standard system phantom for magnetic resonance imaging. Magnetic Resonance in Medicine. 2021;86(3):1194-211.
[7] Hsieh JJ, Svalbe I. Magnetic resonance fingerprinting: from evolution to clinical applications. Journal of Medical Radiation Sciences. 2020;67(4):333-44.
Figures