4343
A Deep Learning-based Multi-Contrast MRI Registration Model with a Realistic Flow Field and Reduced Over-Smoothing Effect1Subtle Medical Inc., Menlo Park, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Data Processing, Registration
To optimize DL-based medical registration models to produce a more realistic flow field and reduce the over-smoothing effect of deformable registration while keeping the generalizability of the multi-contrast registration model to work on all anatomical structures and contrast of MRI, we proposed an algorithm that adopts the image synthetic framework from SynthMorph and optimized with cycle-consistent loss. By experimenting with the jacobian loss, bidirectional loss, and cycle-consistent loss, we managed to further optimize the results of registered images. The evaluation of two MRI image datasets, the BraTS dataset, and the LSpine dataset, demonstrated the increased SSIM, PSNR, and LocalNCC.INTRODUCTION
Medical image registration aligns several medical images based on anatomical structures1. In recent years, deep learning-based registration models have become widely explored. Compared to traditional registration packages like SimpleElastix, DL-based registration models often have quicker inference time and yield competitive or better results[1]. However, particularly in multi-contrast registration, DL-based registration models yield over-smoothed results due to a lack of ground truth training data and image similarity-based training loss. In this study, we propose a new DL-registration model training pipeline that utilizes synthetic images and cycle-consistent loss to deal with the over-smoothing issue which greatly improves the results.METHODS
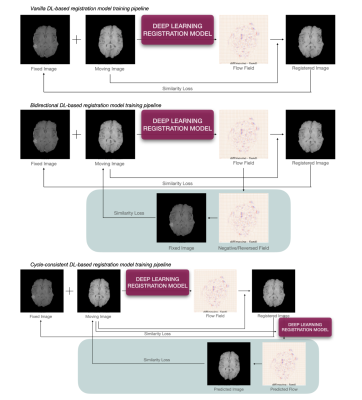
ModelA deep learning 2D image neural network based on UNet, consisting of 4 encodings and 6 decoding convolutional blocks with 128 filters, was used in this study to train the DL-based registration network. The final output of the registration model is a deformation field that is at the same dimension as the input. The models were trained using a combined loss function with an Adam optimizer and StepLR learning rate scheduler for 200 epochs with 256 synthetic label maps and the image pairs they generated through augmentation. Then the models were evaluated using real MRI datasets L-spine and BraTS, with manual random Perlin transformations. Metrics reported include SSIM, PSNR, and LocalNCC.
Data Synthesis
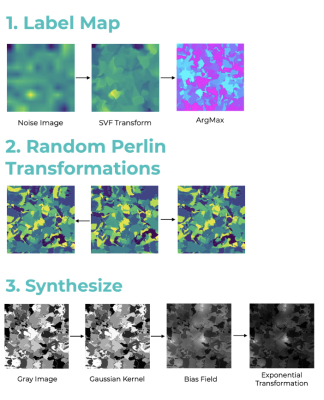
A medical image synthesis pipeline was designed based on SynthMorph[2]. This method generates a label map of random geometric shapes from noise images and a random stationary velocity field (SVF) transforms and assigns labels to the image by selecting the maximum intensity. To generate non-aligned image pairs, a random elastic transformation is applied to get a deformed label map. Then, a gray-scale image is synthesized from a label map by drawing the intensities of the labels from a normal distribution. And the image is further convolved by a gaussian kernel and corrupted by an intensity-bias field. The final image is obtained by min-max scaling and contrast augmentation. Synthetic images are the only images used to train the registration models.
Loss functions
To learn registration, the models used a dice loss to measure the overlapping ratio of labels on the label map after the registration. For the models to yield reasonable images, a LocalNCC loss is used to measure the overall image similarity between the fixed and registered images. Finally, for a less smoothing effect and appropriate flow field, several losses have been applied and compared in this study: the bidirectional loss, the cycle-consistent loss, and the gradient loss.
Datasets
To validate the generalizability and performance of the model on real MRI data, we also included two other datasets in our study: the spine open dataset[3] (464 image pairs) for spine MRI and the BraTS[4] dataset (1251 image pairs) for brain MRI. “L-spine” contains 464 image T1 and T2 image pairs. The BraTS dataset contained 1251 T1 and T2 image pairs. In the evaluation of the registration model, we manually deformed some image pairs using random elastic transformation to get more visible misalignments.
RESULTS
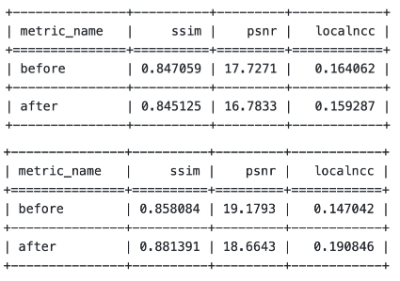
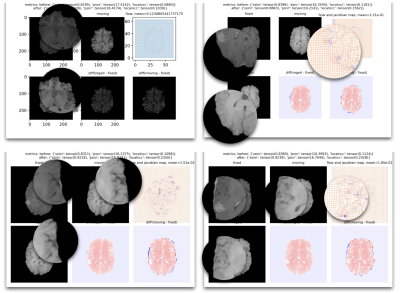
We compared the current four versions of models, all trained on the same synthetic dataset but with different loss combinations. We summarized the results on the BraTS dataset and L-Spine dataset respectively. The results can be summarized by three quantitative metrics: the SSIM, PSNR, and LocalNCC. The quantitative metrics measure the general similarity between the registered image and the fixed image, but cannot fully reflect the registration results. So some examples are also shown.DISCUSSION
Deep learning registration models are more popular in recent years and are known for having better generalizability and quicker inference time. However, traditional registration algorithms and tools like SimpleElastix are still widely used compared to deep learning-based registration models. Most DL registration models are limited in the input anatomy, input image type, contrast, or more generally, image size, etc. And most methods have limited generalizability because it’s hard to collect relevant data for all the contrasts and anatomies with paired registered datasets and the DL models’ performance was capped by the registration quality of the data set collected. The image synthesis pipeline we adopted from SynthMorph solved the problem of data collection and pre-registration. Furthermore, many deep learning registration algorithms, because of weak regularization on the output flow field, often has the issue of an over-smoothing effect because of the overlapped flow field. We experimented with multiple methods and combined useful training losses by modifying the calculation graph. We found that the Jacobian loss with cycle-consistent loss is the best way of dealing with this problem while preserving good registration results.CONCLUSION
In this work, we proposed a new DL-based multi-contrast registration model. The model took advantage of the SynthMorph synthetic pipeline to generate images and thus does need any real medical image pairs. On top of that, we explored several customized loss functions to reduce the over-smoothing effect for the model and got the optimal method using cycle-consistent loss. The proposed model can predict high-quality register images on real datasets L-Spine and BraTS and got significant improvements in SSIM, PSNR, and LocalNCC.Acknowledgements
No acknowledgement found.References
1. Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. Deep learning in medical image registration: a review. Physics in Medicine & Biology. 2020 Oct 7;65(20):20TR01.
2. Hoffmann M, Billot B, Greve DN, Iglesias JE, Fischl B, Dalca AV. SynthMorph: learning contrast-invariant registration without acquired images. arXiv preprint arXiv:2004.10282. 2020 Apr 21.
3. Sudirman, Sud; Al Kafri, Ala; natalia, friska; Meidia, Hira; Afriliana, Nunik; Al-Rashdan, Wasfi; Bashtawi, Mohammad; Al-Jumaily, Mohammed (2019), “Lumbar Spine MRI Dataset”, Mendeley Data, V2, doi: 10.17632/k57fr854j2.2
4. Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging. 2014 Dec 4;34(10):1993-2024.
Figures