4342
Simultaneous Liver and Spleen Segmentation using U-Net Transformer Model on T1-weighted and T2-weighted MRI Data
Huixian Zhang1, Redha Ali1, Hailong Li1, Wen Pan1, Scott B. Reeder2, David T. Harris2, William R. Masch3, Anum Alsam3, Krishna P. Shanbhogue4, Nehal A. Parikh1, Jonathan R. Dillman1, and Lili He1
1Cincinnati Children's Hospital, Cincinnati, OH, United States, 2University of Wisconsin-Madison, Madison, WI, United States, 3Michigan Medicine, University of Michigan, Ann Arbor, MI, United States, 4NYU Langone Health, New York, NY, United States
1Cincinnati Children's Hospital, Cincinnati, OH, United States, 2University of Wisconsin-Madison, Madison, WI, United States, 3Michigan Medicine, University of Michigan, Ann Arbor, MI, United States, 4NYU Langone Health, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
To develop an AI system for precise and fully automated, simultaneous segmentation of the liver and spleen from T1-weighted and T2-weighted MRI data. Our study compares the performance of the U-Net Transformer (UNETR) and the standard 3D U-Net model for simultaneous liver and spleen segmentation using MRI images from pediatric and adult patients from multiple institutions. Our work demonstrates that the UNETR shows a statistical improvement over 3D U-Net in both liver and spleen segmentations.Introduction
Liver and spleen segmentation transforms raw abdominal image data into meaningful, spatially structured information and thus plays an essential role in biomedical image analysis [1]. In the clinical practice, MRI images of different contrasts, such as T1-weighted and T2-weighted images, are commonly acquired to assist diagnosis, treatment planning, and surgery [2]. Hence, segmenting individual liver and spleen structures from MRI T1-weighted and T2-weighted images is an essential ingredient in numerous clinical applications.In this study, we develop a deep-learning-based AI system that is clinically relevant and accurate for fully automatic and simultaneous liver and spleen segmentation from T1-weighted and T2-weighted MRI images. In particular, we employed a U-Net Transformer [3], UNETR, which utilizes a Transformer [4] as the encoder in the U-shaped network (U-Net) architecture [5], to simultaneously segment the liver and spleen.
Methods
Clinical axial T2-weighted fast spin-echo fat-saturated MRI images from three different MRI scanner manufacturers were acquired from four different institutions, on which the liver and spleen were manually delineated by a data analyst and a board-certified radiologist. For T2-weighted imaging, there were 182 patients (age [mean ± standard deviation (SD)], 15.3 ± 3.9 years) from Cincinnati Children’s Hospital Medical Center, 50 patients (age, 51.8 ± 14.3 years) from New York University Medical Center, 62 patients (age, 47.3 ± 15.9 years) from University of Wisconsin, and 47 patients (age, 52.3 ± 14.4 years) from University of Michigan / Michigan Medicine. Clinical axial T1-weighted gradient echo fat-saturated MRI images, also from three different MRI system manufacturers and the same four different institutions, were acquired on which the liver and spleen were manually delineated by a data analyst and a board-certified radiologist as well. For T1-weighted imaging, there were 97 patients (age, 15.8 ± 3.9 years) from Cincinnati Children’s Hospital Medical Center, 49 patients (age, 52.3 ± 14.2 years) from New York University Medical Center, 50 patients (age, 47.9 ± 16.3 years) from University of Wisconsin, and 45 patients (age, 53.3 ± 13.7 years) from University of Michigan / Michigan Medicine.We employed a U-Net Transformer, UNETR, which utilizes a Transformer as the encoder in the U-shaped network (U-Net) architecture to simultaneously and fully automatically segment the liver and spleen. Connecting the transformer encoders to decoders via skip connections at different resolutions enhances the model’s capability for learning long-range dependencies and effectively capturing global contextual representation at multiple scales. The UNETR takes a sequence of sub-volumes with resolution of 32×32×32 sampled from an input MRI image. Each sub volume is parcellated into non-overlapping patches with resolution of 4×4×4 and local self-attentions are calculated to model the patch interactions. We conducted nested random split on the dataset. The Dice similarity coefficient (DICE) was calculated to evaluate the segmentation performance.
Results
Visually, both UNETR and 3D U-Net performed for liver and spleen segmentations that matched closely to our manual segmentations (Figure 1). Quantitatively, as Table 1 indicates, when trained and tested on T2-weighted data only, UNETR achieved a DICE of 0.92±0.07 for liver segmentation and a DICE of 0.91±0.08 for spleen segmentation. This performance was statistically higher (p<0.01) than what 3D U-Net achieved. When trained on T1-weighted data only, UNETR achieved a DICE of 0.93±0.04 for liver segmentation and a DICE of 0.86±0.12 for spleen segmentation. This performance was statistically higher (p<0.01) than what 3D U-Net achieved. The combined T2-weighted and T1-weighted UNETR model achieved a DICE of 0.92±0.08 for liver segmentation on the T2-weighted MRI images and a DICE of 0.92±0.11 for spleen segmentation on the T2-weighted MRI images. It significantly (p<0.01) outperformed 3D U-Net on DICE. At the same time, UNETR model achieved a DICE of 0.93±0.04 for liver segmentation on the T1-weighted MRI images and a DICE of 0.85±0.15 for spleen segmentation on the T1-weighted MRI images. It significantly (p<0.01) outperformed 3D U-Net on DICE. UNETR performed better than 3D U-Net for both organs when trained individually. 3D-Unet suffered from a considerable loss on DICE when trained jointly on T1-weighted and T2-weighted MRI images, however UNETR remained steady on DICE when trained jointly on T1-weighted and T2-weighted MRI images. This phenomenon demonstrates UNETR has better generalizability towards multi-modality MRI data.Conclusions
We have conducted experiments on a large-scale combined pediatric and adult dataset from multi-centers using three MRI system manufacturers, in order to validate the robustness and clinical applicability of our AI system. The UNETR showed a statistical improvement over 3D U-Net in both liver and spleen segmentations and allows simultaneous segmentation of the liver and spleen on both T1-weighted and T2-weighted images.Acknowledgements
This work was supported by the National Institutes of Health [R01-EB030582, R01-EB029944, R01-NS094200, and R01-NS096037]; Academic and Research Committee (ARC) Awards of Cincinnati Children's Hospital Medical Center. The funders played no role in the design, analysis, or presentation of the findings.References
[1] Hollon, T.C., Pandian, B., Adapa, A.R., Urias, E., Save, A.V., Khalsa, S.S.S., Eichberg, D.G., D’Amico, R.S., Farooq, Z.U., Lewis, S. and Petridis, P.D., 2020. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nature medicine, 26(1), pp.52-58.[2] Conte, G.M., Weston, A.D., Vogelsang, D.C., Philbrick, K.A., Cai, J.C., Barbera, M., Sanvito, F., Lachance, D.H., Jenkins, R.B., Tobin, W.O. and Eckel-Passow, J.E., 2021. Generative adversarial networks to synthesize missing T1 and FLAIR MRI sequences for use in a multisequence brain tumor segmentation model. Radiology, 299(2), pp.313-323.
[3] Hatamizadeh, A., Tang, Y., Nath, V., Yang, D., Myronenko, A., Landman, B., Roth, H.R. and Xu, D., 2022. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (pp. 574-584).
[4] Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S. and Uszkoreit, J., 2020. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
[5] Ronneberger, O., Fischer, P. and Brox, T., 2015, October. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
Figures

Figure 1. Qualitative comparisons. a) T1-w of an 18-year-old male subject. U-Net (Dice=0.953 for liver, 0.958 for spleen) UNETR (Dice=0.963 for liver, 0.962 for spleen) b) T1-w of a 16-year-old male subject. U-Net (Dice=0.942 for liver, 0.921 for spleen) UNETR (Dice=0.950 for liver, 0.957 for spleen) c) T2-w of a 15-year-old female subject. U-Net (Dice=0.937 for liver, 0.947 for spleen) UNETR (Dice=0.962 for liver, 0.951 for spleen) d) T2-w of a 17-year-old male subject. U-Net (Dice=0.963 for liver, 0.950 for spleen); UNETR (Dice=0.967 for liver, 0.966 for spleen)
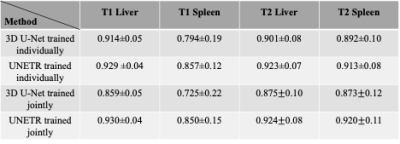
Table 1. Quantitative comparisons between 3D U-Net and UNETR. The first two rows are 3D U-Net and UNETR trained individually on T1-weighted or T2-weighted MRI images. The last two rows are 3D U-Net and UNETR trained jointly on T1-weighted and T2-weighted MRI images at the same time.
DOI: https://doi.org/10.58530/2023/4342