4340
Improving temporal resolution in dynamic hyperpolarized 13C spiral chemical shift imaging using low rank plus local sparse reconstruction1University of Maryland, Baltimore, Baltimore, MD, United States
Synopsis
Keywords: Image Reconstruction, Hyperpolarized MR (Non-Gas)
Improve the accuracy of under-sampled dynamic images for lower intensity metabolites with low rank and local sparsity reconstructionIntroduction
Spiral Chemical Shift Imaging (spCSI) has been an established method for hyperpolarized (HP) 13C MRI with fast acquisition time (Tacq), allowing dynamic imaging with repeated acquisition on the subject over several time frames [1]. In most cases, the spiral readout in k-space is limited by the maximum gradient strength and/or slew rate, requiring multiple interleaves in the spiral k-space trajectory given a specified spectral width. As a result, acquisition of one fully sampled k-space may take a few seconds to complete, leading to a tradeoff between spatial and temporal resolution.Image acceleration for spCSI can be achieved through under-sampling the spiral k-space by acquiring a subset of the full interleaves. Images can be recovered from under-sampled data via low rank (L) plus sparse (S) matrix decomposition model (L+S). In previous study, we have successfully demonstrated the application of L+S reconstruction on in vivo spCSI data. Based on the L+S decomposition algorithm proposed by Otazo et al. [2], we modified the algorithm to solve for low rank component and sparse component of the dynamic images with iterative soft thresholding on the entire spectro-spatio-temporal matrix. In this application, a single threshold is set on the singular values of low rank matrix and another single threshold is set on the sparse-transformed matrix, presented as global low rank and global sparse (GLGS)[3,4]. The algorithm successfully recovers the pyruvate images, but the lactate and alanine images could not be accurately recovered. Since pyruvate contributes to majority of the MR signal, the threshold in the sparsity image is therefore set according to the feature of the dominant component (pyruvate) in the spectro-spatio-temporal matrix, dominating the features of the lower intensity components.
Milshteyn et al. have demonstrated the application of local low rank plus sparse decomposition model for HP 13C MRI with 2D bSSFP sequence (non-spectroscopic imaging)[5]. Spectroscopic imaging features ‘local sparse’ property as neighboring spatial components have similar dynamics after the sparse transform, but each frequency component may not share same dynamics. Hence, we propose a global low rank plus local sparse (GLLS) decomposition for spectroscopic imaging. In GLLS the sparse matrix is divided into multiple blocks along the spectral dimension, and different thresholds are set for each block, thus retaining each frequency component’s dynamic pattern.
Methods
For testing the algorithm, we construct a digital 2D spectroscopic imaging phantom comprising of 4 discs representing the vasculature, kidneys, and liver/body, and 3 spectral peaks (pyruvate, alanine, and lactate), each intensity characterized by the typical dynamic measurement in abdominal region of a mouse following hyperpolarized pyruvate injection. Spiral trajectory with 24 interleaves, 30 x 30 matrix size, 24 echoes and 280 Hz spectral width was used for generation of the ground truth spCSI raw data. Random 8 out of the total 24 interleaves were selected to generate the under-sampled raw data. Difference image are compared between L+S result and ground truth. Normalized root-mean-squared difference (NRMSD) are calculated for the results at each iterations.Two prospectively under-sampled in-vivo imaging experiments were performed on a healthy mouse with injection of hyperpolarized pyruvate. Scan used the same spiral trajectory as in the digital simulation. Scan for the first injection used a constant 5.625° flip angle and the second one used a variable flip angle scheme. The order of the interleaves was randomly permuted for each block. Dynamic images with 3 second temporal resolution can be reconstructed using the fully sampled blocks with conjugate gradient operator (CG, R=1), whereas dynamic images with 1 second temporal resolution can be reconstructed using every 8 interleaves (R=3) via GLGS and GLLS. R=6 was also attempted for the second scan.
Results and Discussion
In simulation, the GLLS result can capture the proper spatial information of the sparse component in those frequency bins, producing a better reconstruction result as seen in the difference image. NRMSD also proves the reduction of error for pyruvate and lactate using GLLS. The residual image Mres at the data coherence step was presented to measure the performance of L+S reconstruction for the in vivo imaging. Mres was significantly reduced using GLLS. Both GLGS and GLLS can successfully restore the pyruvate images without artifacts, but significant distortion of the dynamic curve can be found in lactate and alanine images for GLGS, as seen in Figure 4. Image acceleration effects can be visualized in the vasculature pyruvate for the second scan, where bolus arrival can be determined more accurately at R=3. shown in Figure 5. The results reconstructed with R=6 provides more temporal frames without adding artifacts.Conclusion
GLLS can successfully restore the distinct dynamic pattern of lower intensity metabolites in HP 13C MRI. An effective acceleration of 6 can be achieved using the proposed method without introducing artifacts or distortions.Acknowledgements
This work was supported by NIH grants R21 NS096575, R01 DK106395, R21 CA213020, and R21 CA202694.References
[1] Mayer, Dirk, et al. "Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging." Magnetic resonance in medicine 56.4 (2006): 932-937.
[2] Otazo, Ricardo, et al. "Low‐rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components." Magnetic resonance in medicine 73.3 (2015): 1125-1136.
[3] Zhu, et al. Accelerating Hyperpolarized 13C Spiral Chemical Shift Imaging with Joint Spectral-Spatial Low Rank Plus Sparse Reconstruction, Proc. Intl. Soc. Mag. Reson. Med. 27 (2021)
[4] Zhu, et al. Using Joint Spectral-Spatial Low Rank Plus Sparse (L+S) Reconstruction to accelerate dynamic Hyperpolarized 13C Spiral Chemical Shift Imaging In Vivo, Proc. Intl. Soc. Mag. Reson. Med. 28 (2022).
[5] Milshteyn, Eugene, et al. "Using a local low rank plus sparse reconstruction to accelerate dynamic hyperpolarized 13C imaging using the bSSFP sequence." Journal of Magnetic Resonance 290 (2018): 46-59.
Figures
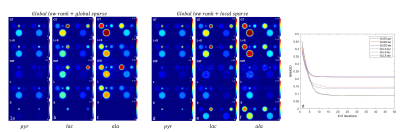
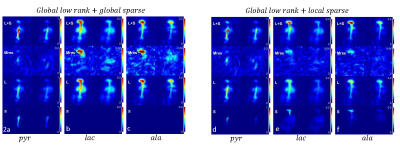
Fig 2a-c, L+S image, residual image (Mres), low rank image (L) and sparse image (S) of the map of spectra after 20 iterations of L+S reconstruction on the in-vivo mouse body imaging using global low rank and global sparsity. a,b,c represents two selected time points of the dynamic images at pyruvate, lactate and alanine frequency bins respectively.
Fig 2d-f, results using L+S reconstruction with global low rank and local sparsity (block size 1 on frequency dimension only).
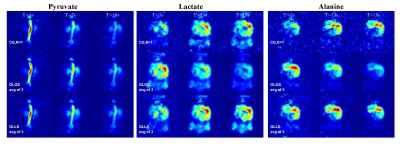
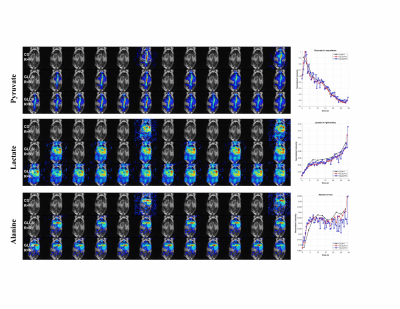
Fig 3 Dynamic images of the 2D coronal mouse body imaging with constant flip angle. Images in each column represents data acquired in a fully sampled block (3 seconds) at specified time.
1st row – R=1 with fully sampled k-space using conjugate gradient operator;
2nd row – average of 3 adjacent map of spectra from the recon result using global low rank + global sparsity
3rd row – average of 3 adjacent map of spectra from the recon result using global low rank + local sparsity
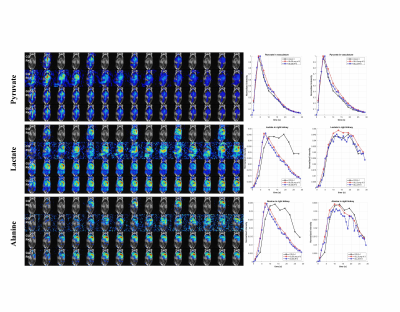
Fig 4 Dynamic images of the coronal mouse body imaging using constant flip angle. T = 7~21s after start of injection
1st row - R=1 with fully sampled k-space at 3s temporal resolution using conjugate gradient operator
2nd row – R=3 with under-sampled k-space at 1s temporal resolution using conjugate gradient operator; (result of L+S reconstruction at iteration 0)
3rd row – R=3, L+S reconstruction with global low rank and global sparsity
4th row – R=3, L+S reconstruction with global low rank and local sparsity.
Dynamic curves are plotted for the selected ROI (white rectangle)