4327
Data Standardization and DICOM Integration for Hyperpolarized 13C MRI1Radiology & Biomedical Images, University of California,San Francisco, San Francisco, CA, United States
Synopsis
Keywords: Hyperpolarized MR (Non-Gas), Contrast Mechanisms, Data,DICOM
Hyperpolarized (HP) 13C MRI has shown promise as a valuable modality for in vivo measurements of metabolism, and is currently in human trials at over 15 human research sites worldwide. It is important to adopt standardized data storage practices as it will allow sites to meaningfully compare data. We propose using specific Attributes in DICOM format, including “Contrast Agents”, and provide a Python based implementation to add these values for individual studies. Moreover, we propose best practices for HP 13C MRI data storage that will support future multi-site trials and research studies and technical developments of this imaging technique.
Introduction
Hyperpolarized (HP) 13C MRI has shown promise as a valuable modality particularly for in vivo measurements of metabolism1, and is currently in human trials at over 15 human research sites worldwide with 73+ human research publications. It is based on intravenous injection of a hyperpolarized contrast agent that has been enriched with 13C. The most widely used agent is 13C-pyruvate for measuring metabolism, but there are also a range of other promising agents undergoing clinical translation including 13C-urea (perfusion), 13C-alpha-ketoglutarate (metabolism), and 13C-fumarate (necrosis).As HP 13C MRI continues to grow, it is important to adopt standardized data storage practices as it will allow sites to meaningfully compare data. The DICOM format is an attractive standardization format as it is commonly used in digital imaging, medical imaging, and communications. The DICOM file type consists of a header and image data sets packed into a single file. The information in the header is organized as a constant and is standardized by a series of tags. By extracting the data from these tags, we can access important metadata regarding the patient demographics and study parameters that are crucial for study interpretation. We propose to use the “Contrast Agents Attributes” of the DICOM format to store HP 13C agent specific information and incorporate these details into the header using a Python script. Moreover, we propose best practices for HP 13C MRI data storage that will support future multi-site trials and research studies and technical developments of this imaging technique.
Methods
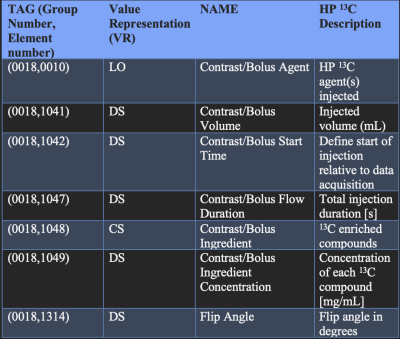
Our current processing pipelines support DICOM creation from multiple vendors and pulse sequences, listed in Figure 1. The spectroscopy sequences use SIVIC for DICOM creation which includes support for DICOM Spectroscopy2,3.We are using the tool PyDicom to access the data in DICOM files. PyDicom is a general-purpose DICOM framework whose purpose is to read and write DICOM attributes. We can also add, delete, and modify any attributes to DICOM images. A DICOM attribute consists of a tag used for identification which is comprised of hexadecimal numbers denoting a DICOM Group Number and DICOM Element Number. There is also a DICOM Value representation (VR) that represent the type of data and the format of the attribute value.
Currently, the HP 13C images created by MRI scanners or in-house reconstruction pipelines do not utilize the Contrast/Bolus Attributes. We seek to leverage the existing Contrast/Bolus Attributes in DICOM Images to store information to identify the study and experimental parameters that are valuable for HP 13C data analysis by researchers and clinicians. Figure 2 lists the attributes we are adding to our images. For metabolite-specific imaging, we have also included the metabolite-specific flip angles in the DICOM header, as these were not captured by default during the DICOM creation.
Results
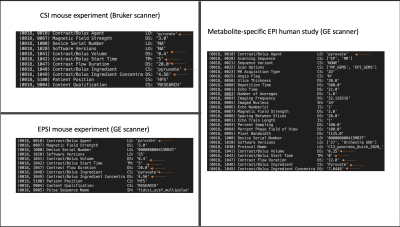
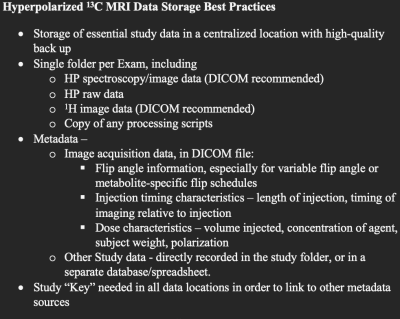
The script utilizing PyDicom was successfully able to add all the attributes to the images as shown in the example in Figure 3. These figures show example values for the DICOM attributes in Figure 2. This can be applied in batch mode to many DICOM files as well.In addition to these specific DICOM attributes, we have created a proposed Data Storage Best Practices in Figure 4. These were based upon a survey of research groups at our institution and are being implemented across new studies.
Discussion and Conclusion
We have proposed a set of DICOM attributes to store HP 13C MRI metadata, particularly regarding the HP agent, and implemented via PyDicom a program to add these attributes based on user input. In addition, we have proposed a broader set of data storage best practices. These are currently undergoing implementation at our institution, and we look forward to receiving further feedback from the HP 13C MRI community on this effort. The efforts and results of the HP 13C MRI Consensus Group that was formed in 2022 will provide an excellent way to discuss and finalize these data standardization effortsAcknowledgements
This work was supported by NIH Grants R01CA249909 & P41EB013598References
[1] Hyperpolarized 13C MRI: State of the Art and Future Directions. Radiology. 2019 05; 291(2):273-284. Wang ZJ, Ohliger MA, Larson PEZ, Gordon JW, Bok RA, Slater J, Villanueva-Meyer JE, Hess CP, Kurhanewicz J, Vigneron DB. PMID: 30835184; PMCID: PMC6490043.
[2]SIVIC.Available online at: https://sourceforge.net/p/sivic/sivicwiki/Home/ DOI: 10.5281/zenodo.4777197
[3] Crane, Jason C., Marram P. Olson, and Sarah J. Nelson. “SIVIC: Open-Source, Standards-Based Software for DICOM MR Spectroscopy Workflows.” International Journal of Biomedical Imaging 2013 (July 18, 2013): e169526. https://doi.org/10.1155/2013/169526.
[4] Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. J Magn Reson. 2010 Jul; 205(1):141-7. Wilson DM, Keshari KR, Larson PE, Chen AP, Hu S, Van Criekinge M, Bok R, Nelson SJ, Macdonald JM, Vigneron DB, Kurhanewicz J. PMID: 20478721; PMCID: PMC2885774.
[5] Development of specialized magnetic resonance acquisition techniques for human hyperpolarized [13 C,15 N2 ]urea + [1-13 C]pyruvate simultaneous perfusion and metabolic imaging. Magn Reson Med. 2022 09; 88(3):1039-1054. Liu X, Tang S, Mu C, Qin H, Cui D, Lai YC, Riselli AM, Delos Santos R, Carvajal L, Gebrezgiabhier D, Bok RA, Chen HY, Flavell RR, Gordon JW, Vigneron DB, Kurhanewicz J, Larson PEZ. PMID: 35526263.
[6] Development of a symmetric echo planar imaging framework for clinical translation of rapid dynamic hyperpolarized 13 C imaging. Magn Reson Med. 2017 02; 77(2):826-832. Gordon JW, Vigneron DB, Larson PE. PMID: 26898849; PMCID: PMC4992668
Figures