4324
Bias Correction for Improved Quantification of Hyperpolarized [1-13C]Pyruvate Cerebral Perfusion1UC San Francisco, San Francisco, CA, United States, 2MR Research Center, Clinical Medicine, Aarhus University, Aarhus, Denmark
Synopsis
Keywords: Hyperpolarized MR (Non-Gas), Perfusion, Brain
Bias correction of hyperpolarized [1-13C]pyruvate brain MRI allows for improved quantification of cerebral perfusion. In the healthy volunteers studied, bias corrected pyruvate and ASL rCBF exhibited higher correlation than uncorrected pyruvate and ASL rCBF. We hypothesize that N4ITK bias correction can be used to remove low frequency inhomogeneities from hyperpolarized MRI before single-metabolite analyses.
Introduction
Hyperpolarized (HP) 13C pyruvate MRI is a valuable metabolic imaging technique1 that can also be applied to the assessment of perfusion2, as previously shown using multi-resolution acquisitions3. When multichannel receiver arrays are utilized in HP studies to improve SNR and increase spatial coverage, they introduce coil weighting which is challenging to remove due to the difficulty in estimating coil sensitivity profiles in HP 13C experiments4. As an alternative to directly measuring the coil sensitivity, bias correction algorithms can remove the low frequency coil shading and have been successfully applied to multiple MR image acquisition strategies including hyperpolarized 3He gas lung images5. Building upon our prior work where we compared 1H ASL to 13C pyruvate perfusion values2 in the brain, this study investigated N4ITK bias correction for improving the quantification of cerebral perfusion in HP pyruvate MRI studies.Methods
HP [1-13C]pyruvate brain MRI examinations of 5 healthy subjects (7 total studies) were acquired with a multi-resolution metabolite-selective EPI acquisition3. A volume transmit 13C coil was used for RF excitation in combination with a dual-tuned 1H/13C 8/24-channel array for reception (RAPID Biomedical, Germany) on a 3T scanner (MR750, GE Healthcare). In-plane resolution was 7.5x7.5 mm2 for pyruvate and 15x15 mm2 for lactate and bicarbonate. The slice thickness was 15 mm for all metabolites. The HP MRI scan time was one minute, including 20 time points with a 3-s temporal resolution. HP data was reconstructed using methods described in Gordon et al3 and denoised using patch-based higher-order singular value decomposition6. For anatomic reference and proton perfusion, 3D T1 IR-SPGR, T2 FSE, and 3D ASL datasets were acquired.Bias correction using N4ITK5 was performed on HP 13C and ASL images. To obtain 13C bias fields, denoised lactate area-under-curve (AUC) images were brain-masked and input to N4ITK using 3 layers of 100 iterations, convergence threshold of 0 to run all iterations, B-spline order of 3, and a shrink factor of 1. The resulting bias field was then used to correct pyruvate dynamic images before perfusion calculation. ASL images were also bias corrected with the same parameters because there was noticeable asymmetry on the left versus right side of the brain.
Bias corrected dynamic pyruvate images were used to obtain pyruvate perfusion parameters. An arterial input function was fitted using up to four voxels in each study7. Relative CBF (rCBF) was calculated using block-circulant singular value decomposition and corrected for depolarization8. Voxels with pyruvate rCBF >0.3 were excluded from correlation because they were within the superior sagittal sinus. ASL perfusion maps were resampled to the pyruvate voxel size using tri-linear interpolation. Gray and white matter masks were segmented on T2 FSE images, then cortex was excluded from white matter masks using the Automated Anatomical Atlas 39. Voxels with at least 90% brain, 50% gray matter, or 50% white matter were included in the correlation analysis. Pearson correlation coefficients were calculated for 1H ASL and 13C pyruvate rCBF.
Results & Discussion
A visual summary of HP 13C MRI bias correction is shown in Figure 1, with brain-masked lactate AUC images used as the input to the algorithm. Pyruvate AUC images were not used to estimate the bias field because of the heavy weighting from large pyruvate signals in the anterior and posterior portions of the superior sagittal sinus (Figure 2). Using the lactate-derived 13C bias fields to correct pyruvate images before perfusion calculation decreased the intensity weighting most pronounced at the edges of the brain due to coil proximity. The bias fields generated by lactate AUC images were qualitatively similar to the expected coil profiles and were visually consistent across the 5 subjects shown in Figure 3.Bias-corrected pyruvate and ASL perfusion maps were compared using a normalized rCBF difference map shown in Figure 4. Both ASL and pyruvate rCBF maps demonstrated higher perfusion in the gray matter than in the white matter, but normalized ASL rCBF was approximately 40% higher than pyruvate rCBF in gray matter for the subject shown. In the white matter, the normalized rCBF difference was about 30%. These trends are continued for the correlations between ASL and pyruvate rCBF for all subjects shown in Figure 5. The correlation coefficients for bias-corrected brain, gray matter, and white matter voxels were 0.58, 0.55, and 0.76 respectively (all p-values < 10-4). These positive correlations reflect the same patterns as our prior pyruvate perfusion work2, but all the correlations from bias-corrected perfusion maps were greater than those from uncorrected perfusion maps (Figure 5, top row). The largest increase in correlation was for gray matter voxels due to the correction of coil shading at the edges of the brain.
Conclusion
Bias correction removed coil shading from HP MRI without directly measuring sensitivity profiles and increased correlation between pyruvate and ASL perfusion values. Low frequency intensity variations such as coil shading and field inhomogeneity can impact quantification of HP images using single-metabolite methods such as z-scores, unnormalized AUC, and perfusion fitting. The N4ITK bias correction used in this study can be applied to metabolite images before further analysis and can improve quantification of HP MRI.Acknowledgements
This research was supported by NIH grants P01CA118816, U01EB026412, P41EB013598, and the UCSF NICO project.References
1. Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Sci Transl Med. 2013;5(198):198ra108. doi:10.1126/scitranslmed.30060702. Hu JY, Vaziri S, Bøgh N, et al. Investigating Cerebral Perfusion with High Resolution Hyperpolarized [1-13C]Pyruvate MRI. ISMRM abstract and poster. 2022 May.
3. Gordon JW, Autry AW, Tang S, et al. A variable resolution approach for improved acquisition of hyperpolarized 13C metabolic MRI. Magn Reson Med. 2020;84(6):2943-2952. doi:10.1002/mrm.28421
4. Hansen RB, Sánchez-Heredia JD, Bøgh N, et al. Coil profile estimation strategies for parallel imaging with hyperpolarized 13C MRI. Magn Reson Med. 2019;82(6):2104-2117.
5. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310-1320. doi:10.1109/TMI.2010.2046908
6. Kim Y, Chen HY, Autry AW, et al. Denoising of hyperpolarized 13C MR images of the human brain using patch-based higher-order singular value decomposition. Magn Reson Med. 2021;86(5):2497-2511. doi:10.1002/mrm.28887
7. Peruzzo D, Bertoldo A, Zanderigo F, Cobelli C. Automatic selection of arterial input function on dynamic contrast-enhanced MR images. Comput Methods Programs Biomed. 2011;104(3):e148-e157. doi:10.1016/j.cmpb.2011.02.012
8. Johansson E, Månsson S, Wirestam R, et al. Cerebral perfusion assessment by bolus tracking using hyperpolarized 13C. Magn Reson Med. 2004;51(3):464-472. doi:10.1002/mrm.20013
9. Rolls ET, Huang CC, Lin CP, Feng J, Joliot M. Automated anatomical labelling atlas 3. Neuroimage. 2020;206:116189. doi:10.1016/j.neuroimage.2019.116189
Figures
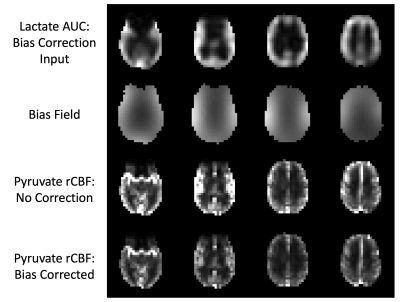
Figure 1. Visual summary of hyperpolarized 13C MRI bias correction. Lactate area-under-curve (AUC) images are used as the N4ITK bias correction input because pyruvate images have extremely high vascular signals that are not reflective of the underlying coil profile. The resulting bias field is then applied to pyruvate images before calculating relative cerebral blood flow (rCBF).
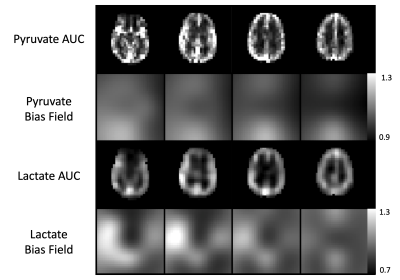
Figure 2. Comparison of bias fields generated from pyruvate area-under-curve (AUC) versus lactate AUC. The pyruvate bias field is weighted primarily in the anterior and posterior portions of the superior sagittal sinus. The lactate bias field is qualitatively similar to the expected coil profile with weighting around the edges of the brain.
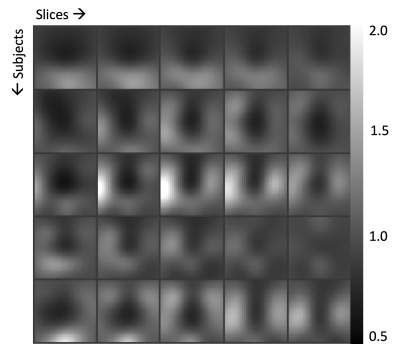
Figure 3. Bias fields of hyperpolarized 13C brain MRI from 5 subjects. Bias fields were generated using the N4ITK algorithm applied to lactate AUC images and were consistent across all studies.
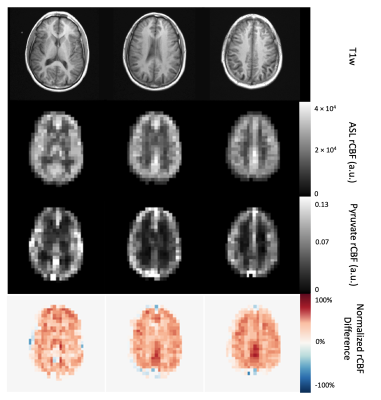
Figure 4. ASL perfusion maps resampled to the [1-13C]pyruvate resolution (7.5 x 7.5 x 15 mm3) and [1-13C]pyruvate rCBF in a normal human brain with T1-weighted anatomical reference MRI and normalized rCBF difference map. Both ASL and pyruvate rCBF maps demonstrated higher values in the gray matter than in the white matter, but the normalized ASL rCBF was approximately 40% higher than normalized pyruvate rCBF in gray matter and 30% higher in white matter. rCBF is in arbitrary units.
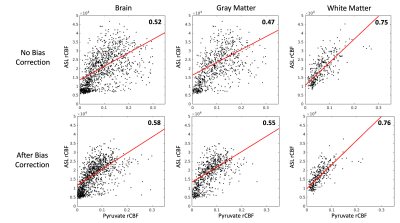
Figure 5. Pyruvate rCBF versus ASL rCBF linear correlation coefficients and scatterplots for voxels from all subjects (n = 5) and all studies (n = 7), by respective mask (5349 brain, 1806 gray matter, and 946 white matter voxels). The top row are rCBF values before bias correction, and the bottom row are rCBF values after bias correction. The apparent spread of rCBF values is reduced and correlation is increased after bias correction for ASL and pyruvate. All units are arbitrary, and all correlation p-values were less than 10-4.