4298
Fast reconstruction of SMS bSSFP myocardial perfusion images using a combination of parallel imaging and AI-based denoising1School of Biomedical Engineering and Imaging Sciences, Faculty of Life Sciences and Medicine, King's College London, London, United Kingdom, 2MR Research Collaborations, Siemens Healthcare Limited, Camberley, United Kingdom, 3Cardiovascular Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 4MR Research Collaborations, Siemens Healthcare Limited, Melbourne, Australia
Synopsis
Keywords: Heart, Perfusion
Simultaneous multi-slice (SMS) imaging enables high spatial coverage and resolution for myocardial perfusion imaging. We developed and evaluated a fast, noise-removing and signal-preserving SMS reconstruction approach combining parallel imaging and deep-learning and evaluated its benefit for myocardial perfusion imaging. In comparison to conventional parallel imaging reconstruction, the proposed SMS reconstruction provides improved perceived SNR and image quality (p<0.008), preserved sharpness and signal fidelity (p>0.05) in a short computation time (+20s for an entire dataset).Introduction
Stress first-pass myocardial perfusion imaging is widely used clinically for the assessment of suspected coronary artery disease. Simultaneous multi-slice (SMS) is an acceleration technique which has been successfully combined with bSSFP imaging and iterative reconstruction (Iter) for myocardial perfusion with high spatial resolution, increased spatial coverage, improved image quality, and high diagnostic accuracy 1,2. However, iterative reconstruction can cause undesirable signal infidelity due to spatio-temporal regularization. It is also computationally expensive, limiting its online use. In this study, we present a fast denoising and signal-preserving reconstruction technique for SMS-bSSFP myocardial perfusion imaging that combines parallel imaging and AI.Methods
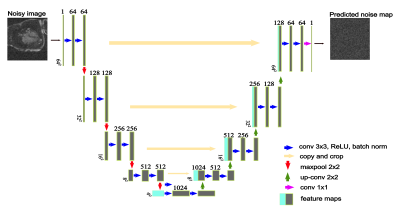
Proposed SMS-bSSFP Acquisition and ReconstructionSMS-bSSFP was implemented with RF phase-based CAIPIRINHA encoding 3, GC-LOLA and a “lean” implementation for slice separation 4. Images were reconstructed using TGRAPPA, followed by non-rigid image registration and AI-based image denoising using a proposed 2D noise map estimating Unet (NoiseMapNET), as described in the next section.
NoiseMapNET Architecture
Figure 1 shows the 2D NoiseMapNET, which takes as an input a noisy image and outputs its predicted residual noise map. The model learned by minimizing the mean square error (L2) loss function of the predicted residual noise map to the ground truth generated noise map. The model parameters were: learning rate=1e-5, early stopping, dropout rate=0.5, ReLU activation, batch normalization, and an Adam optimizer (𝛽1=0.9, 𝛽2=0.999). The denoised image was obtained by subtracting the predicted residual noise map from the input noisy image.
The model was trained using 300 high SNR four-chamber CINE images split into training (80%) and validation (20%) sets. Preprocessing included patch-based learning (64x64), gaussian noise addition (equivalent to blood SNR of ~10) and standardization. Training of the model was implemented using Python language and Pytorch framework and required approximately 4 hours on a PC with GPU (NVIDIA Titan V).
Experimental Evaluation
Seventeen patients (60±15 years), with suspected coronary artery diseases were recruited for a stress perfusion scan using a research application SMS bSSFP sequence on a 1.5T scanner (Magnetom Aera, Siemens Healthcare, Erlangen, Germany). Imaging parameters were: 6 slices, FOV 360x360mm2, resolution 1.9x1.9mm2, slice thickness 10mm, TR/TE 2.9ms/1.24ms, flip angle 50°, 80 dynamics, effective saturation recovery time 94s, bandwidth 1302Hz/px, in-plane acceleration factor 2.5, multiband factor 2, TGRAPPA undersampling. Images were reconstructed using: 1) Iterative reconstruction with integrated motion correction 5 (Iter), 2) TGRAPPA followed by non-rigid image registration and 3) proposed reconstruction.
Quantitative and Qualitative Analysis
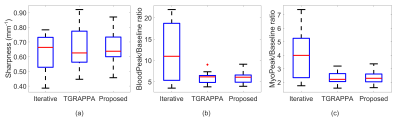
Quantitative and qualitative analysis was performed to compare the three reconstruction approaches. Quantitative myocardial sharpness index at the myocardium and blood boundary for the dynamic corresponding to peak blood signal was measured as previously described 5. Additionally, signal ratios of peak-to-baseline myocardium signal (PBmyo), and peak-to-baseline left ventricle (LV) blood pool signal (PBblood), were calculated to evaluate signal consistency of the reconstruction techniques. For qualitative analysis, two expert clinicians blinded from the clinical and reconstruction information consensually scored the images from the three reconstruction techniques. Metrics evaluated were perceived SNR (pSNR) (0-poor; 3-high), image quality (0-poor; 3-excellent) and number of diagnostic myocardial segments.
Results
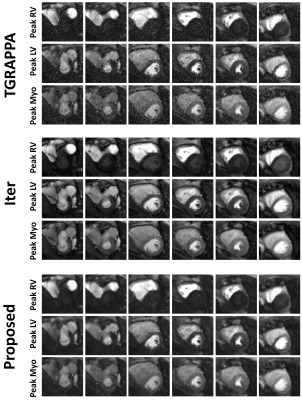
Figures 2 and 3 show example reconstructions and corresponding predicted noise maps from the same subject, respectively. Over all patients, the Iter method led to the highest pSNR (3.0±0.0 vs. 2.0±0.0 (proposed) and 1.3±0.6 (TGRAPPA), p<0.001 for both) and image quality (2.7±0.4 vs. 1.8±0.4 (proposed) and 1.3±0.6 (TGRAPPA), p<0.001 for both). However, it also significantly changed the PBmyo/PBblood ratios (3.9±1.7/11.8±7.0 vs. 2.3±0.5/5.8±1.3 (proposed) and 2.3±0.5/5.8±1.3 (TGRAPPA, used as the reference), p<0.001 for both) and had much longer computation time (12min vs. 20s (proposed) for an entire dataset).In comparison to TGRAPPA, the proposed reconstruction had higher image quality (p=0.008), pSNR (p<0.001) without any significant changes to the PBmyo /PBblood ratios (p=0.89/0.53). There were no significant differences in terms of average myocardial sharpness (0.63±0.1mm-1 (Iter), 0.65±0.1mm-1 (proposed), 0.65±0.1mm-1 (TGRAPPA), p=0.77) and rate of diagnostic segments between all techniques (100±0% (Iter), 100±0% (proposed), 94.1±23.5% (TGRAPPA), p=0.36) (Figures 4, 5).
Discussion
The proposed reconstruction improved image quality and pSNR, preserved signal fidelity/sharpness in the images, and only required minimal additional computational time, compared to TGRAPPA. Therefore, it may be valuable for online image reconstruction and accurate perfusion quantification. Iter resulted in bias in PBmyo/PBblood ratios, likely caused by the spatial-temporal regularization 6, and had a long computation time, limiting its applicability for online reconstruction at present. However, Iter also provided the highest image quality and pSNR, which may be valuable for retrospective image assessment. Confounding factors to perfusion defects such as dark rim/motion artifacts could also have their temporal profile altered/smoothed temporally using Iter, making their identification more complicated. The proposed reconstruction also has potential to facilitate the detection of such artefacts. There may be value in using both the proposed reconstruction and Iter concomitantly as they may provide complementary information. Future studies in a larger cohort of patients is now warranted.Conclusion
A fast and signal/feature preserving AI-based reconstruction of SMS perfusion images resulted in improved image quality and pSNR, with respect to standard parallel imaging techniques. This approach may offer a valuable alternative for online SMS bSSFP reconstruction and accurate myocardial quantification, and could be used concomitantly to iterative reconstruction.Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) grants (EP/R010935/1), the Centre for Doctoral Training by King’s College London’s Centre for Doctoral Studies, the British Heart Foundation (BHF) grants (PG/19/11/34243 and PG/21/10539), the Wellcome EPSRC Centre for Medical Engineering at King’s College London (WT 203148/Z/16/Z), the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ National Health Service (NHS) Foundation Trust, King’s College London and the Botswana government. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.References
1. Nazir MS, Neji R, Speier P, et al. Simultaneous multi slice (SMS) balanced steady state free precession first-pass myocardial perfusion cardiovascular magnetic resonance with iterative reconstruction at 1.5 T. Journal of Cardiovascular Magnetic Resonance. 2018;20(1).
2. Nazir Muhummad S, Milidonis X, McElroy S, et al. Quantitative Myocardial Perfusion With Simultaneous-Multislice Stress CMR for Detection of Significant Coronary Artery Disease. JACC: Cardiovascular Imaging. 2022;15(9):1672-4.
3. Stäb D, Ritter CO, Breuer FA, et al. CAIPIRINHA accelerated SSFP imaging. Magnetic Resonance in Medicine. 2011;65(1):157-64.
4. Stäb D, and Speier P. Gradient-controlled local Larmor adjustment (GC-LOLA) for simultaneous multislice bSSFP imaging with improved banding behavior. Magnetic Resonance in Medicine. 2019;81(1):129-39.
5. Mcelroy S, Kunze KP, Nazir MS, et al. Simultaneous multi-slice steady-state free precession myocardial perfusion with iterative reconstruction and integrated motion compensation. European Journal of Radiology. 2022;151:110286.
6. Jaspan ON, Fleysher R, and Lipton ML. Compressed sensing MRI: a review of the clinical literature. The British journal of radiology. 2015;88(1056):20150487.
Figures