4228
Automatic Alignment of Ex-vivo Brain Pathology to 7T structural MRI
Jinghang Li1, Nadim Farhat1, Jacob P. Berardinelli1, Joseph M. Mettenburg2, Howard J. Aizenstein1,3, Julia K. Kofler4, and Tamer S. Ibrahim1,3
1Bioengineering, University of Pittsburgh, Pittsburgh, PA, United States, 2Neuroradiology, University of Pittsburgh Medical Center, Pittsburgh, PA, United States, 3Psychiatry, University of Pittsburgh, Pittsburgh, PA, United States, 4Pathology, University of Pittsburgh Medical Center, Pittsburgh, PA, United States
1Bioengineering, University of Pittsburgh, Pittsburgh, PA, United States, 2Neuroradiology, University of Pittsburgh Medical Center, Pittsburgh, PA, United States, 3Psychiatry, University of Pittsburgh, Pittsburgh, PA, United States, 4Pathology, University of Pittsburgh Medical Center, Pittsburgh, PA, United States
Synopsis
Keywords: High-Field MRI, Ex-Vivo Applications
In this work, we present an image alignment pipeline that facilitates ongoing Alzheimer’s disease (AD) white matter pathology studies. Following our recently developed ex-vivo 7T cutting-guide container [1], in this work, we demonstrate a standardized alignment (between histopathology and ex-vivo 7T MRI) procedure that significantly reduces human effort and eliminate the need for manual alignment.Introduction
7 tesla (T) magnetic resonance imaging (MRI) offers substantial fine details at in-vivo neuro imaging. Further, with extended scan time, zero motion artifacts and zero cardiovascular movements, 7T MRI can bring the ex-vivo scan resolution to a level that is unparallel [1] [2] . Pinpoint white matter lesion examination coupled with 7T MRI ex-vivo scans opens the doors for precise AD pathology studies. Our previous work using an ex-vivo 7T cutting-guide container has allowed precise cutting and offered reference points for easier visual alignment [1]. The image alignment is crucial in postmortem AD pathology studies because white matter lesions are not visible on pathology camera images, thus relying on the alignment of structural ex-vivo MRI to locate the white matter lesions (Figure 1).However, this visual alignment between the T1-w MR images and the camera projection images is often tedious and time consuming. To reduce the manual effort involved in the alignment process, we propose a pipeline that significantly reduces the human labor.Methods
The alignment pipeline proposed in this work is shown in Figure 2. All the camera images are the coronal view of the sliced postmortem brain. The image background was first removed using the open-source project “rembg”. Later, the processed images underwent the histogram-based intensity normalization to make pathology camera images look more like the T1-w MR images [3]. T1-w MRI histogram was used because of the tissue contrast resemblance to that of the ex-vivo pathology. After all the image preprocessing steps are done, the T1-w volume matrix is then rotated in the sagittal dimension (see Figure 2). At each rotation angle, every coronal MRI slice in the rotated matrix was compared with all the processed ex-vivo pathology camera image using structural similarity index [4] resulting in the subsequent curve shown in Figure 2. At each panel, the highest similarity index location indicates the matching slice location. To further improve the matching accuracy, we added position gates to the similarity index curves, restricting the matching slice indices to be within the corresponding range.After finding all the matching coronal MRI slices, the rotation angles were saved to apply the transformation matrix on the co-registered T2-weighted MR image to extract the corresponding T2-w tissue contrast for all the matching slices.
Results
Figure 3 shows the preliminary matched T1-w MR images before the position gates. As it can be seen in the last column, the algorithm struggles to find the correct matching slices without the position gates. Figure 1 shows the matched T1-w and T2-w MR images after the position gates. We have successfully applied this pipeline on 9 ex-vivo brain scans.Discussion and Conclusions
In this work, we presented a preliminary ex-vivo brain pathology to T1-w MR image alignment pipeline that significantly reduces human effort by avoiding manual alignment. The pipeline leverages various image processing techniques. While computing structural similarity index can help locate most of the matching slices, interventions are still needed for some edge cases. One remediation we implemented in this work is by applying the position gates, thus restricting the matching slice indices to be within the corresponding range.Acknowledgements
This work was supported by the National Institutes of Health under award number: R01AG067018, RF1AG025516, R01MH111265, R01AG063525, R56AG074467.References
1.Berardinelli, J., et al. Reusable 3D Printed Ex-vivo Brain Enclosure and Two-Piece Cutting Guide for Axial and Coronal MRI Registration with Gross Anatomy Photographs. in International Society of Magnetic Resonance in Medicine Annual Meeting. 2021. London, UK.
2.Farhat N., e.a. Addressing ultra highfield MRI challenges in ex-vivo. in International Society of Magnetic Resonance in Medicine Annual Meeting. 2021. London, UK.
3.Nyúl, L.G. and J.K. Udupa, On standardizing the MR image intensity scale. Magn Reson Med, 1999. 42(6): p. 1072-81.
4.Zhou, W., et al., Image quality assessment: from error visibility to structural similarity. IEEE Transactions on Image Processing, 2004. 13(4): p. 600-612.
Figures
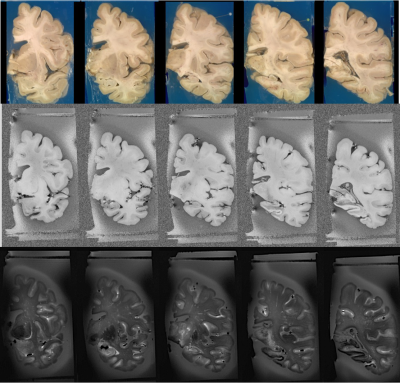
Figure 1 Matching coronal slices in the T1-w and T2-w MR images after position gates.

Figure 2 Diagram of the automatic alignment workflow. Open-source project rembg was used to remove the background, and histogram-based intensity normalization was used before comparing with the rotated MRI images.
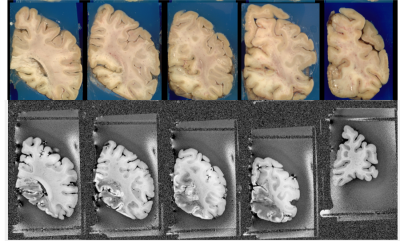
Figure 3 Matched T1-w MR images before position gates.
DOI: https://doi.org/10.58530/2023/4228