4205
Patient-specific Self-supervised Resolution-enhancing Networks for Synthesizing High-resolution Magnetic Resonance Images
Xiaofeng Yang1, Sagar Mandava2, Yang Lei1, Huiqiao Xie1, Tonghe Wang3, Justin Roper1, Tian Liu4, and Hui Mao1
1Emory University, Atlanta, GA, United States, 2GE Healthcare, Atlanta, GA, United States, 3Memorial Sloan Kettering Cancer Center, New York, NY, United States, 4Icahn School of Medicine at Mount Sinai, New York, NY, United States
1Emory University, Atlanta, GA, United States, 2GE Healthcare, Atlanta, GA, United States, 3Memorial Sloan Kettering Cancer Center, New York, NY, United States, 4Icahn School of Medicine at Mount Sinai, New York, NY, United States
Synopsis
Keywords: Quantitative Imaging, Data Processing
This study aims to develop an efficient and clinically applicable method using patient-specific self-supervised resolution-enhancing network to synthesize the high-resolution information of MR images in the low-resolution direction to generate respective high-resolution MRI.Introduction
High-resolution (HR) images are desirable, sometimes essential, in detecting and characterizing subtle differences in structural abnormalities and small malignant lesions, e.g., < 3-5 mm. However, acquiring HR images with less than 0.5 mm normally comes with a lower signal-to-noise ratio (SNR) and contrast when altering repetition time1, prolonged scan time, and subsequently, high risk for motion artifacts. It is even challenging for current clinical scanners with the standard gradient power to obtain high through-plane resolution (or slice direction), especially for the large FOV required in abdominal/pelvic scans that need to have enough coverage and avoid aliasing2. Thus, the diagnostic accuracy and fidelity of lesion segmentation and quantification are then compromised by the low resolution in the through-plane direction, especially when the targeted or suspicious abnormalities are less than 5 mm. Therefore, it is essential to develop an efficient and clinically applicable method to predict the HR information of the MR image in the low-resolution (LR) direction (e.g., 3 mm thick slice in pelvis images) to generate respective HR images (e.g., 1 mm).Methods
We report a self-supervised deep learning algorithm to enhance image resolution, which does not use any training images with a high through-plane resolution, yet can still generate high through-plane resolution images only relying on the acquired images with low through-plane resolution. The workflow consists of training two deep neural networks in parallel that independently predict the HR images in x- and y- planes (i.e., sagittal and coronal planes) from images acquired in the axial direction, respectively. Then the final synthesized HR MR images are generated by fusing the two predicted images. In each of the parallel network paths, the first step is to formulate the training data by resampling the original LR images along the x-axis (or y-axis) in the latitudinal plane. Image patches are then extracted to build the paired training dataset with high and low resolution. The paired patches are input into the networks to derive end-to-end non-linear mapping. Then the final synthetic HR images are generated by fusing the two predicted images. We used 250 brain patient MR images obtained from the multimodal BraTS2020 dataset to evaluate the proposed workflow. HR images were generated in the axial, sagittal, and coronal directions. In each direction, ground truth images were down-sampled by a factor of 3 to simulate low through-plane MR images to test our model. Normalized mean absolute error (NMAE) and peak SNR (PSNR) were used to quantify the performance. MR images (20 patients) collected from a GE 3T PET/MRI scanner were used for a proof-of-concept study of the proposed method since no HR images are available as the ground truth.Results
Quantitative evaluations of the synthetic HR MR images from the brain MR dataset show that image quality is enhanced for T1, contrast-enhanced T1 (T1CE), T2, and T2 FLAIR images. The mean NMAE and PSNR were 0.053±0.017 and 34.1±1.7 dB for T1, 0.069±0.025 and 31.3±2.0 dB for T1CE, and 0.068±0.026 and 31.2±1.8 dB for T2, as well as 0.051±0.018 and 34.5±2.1 dB for FLAIR images, demonstrating the accuracy of the proposed method. Figure 1 provides a comparison of native LR (0.98×0.98×6.54 mm3) and enhanced HR images of a pelvis study from the clinical site protocol. With bi-cubic interpolation, the clarity of the synthesized sagittal and coronal images is improved; however, even better edge definition and sharper textures are achieved in the sagittal and coronal images using the reported self-supervised resolution enhancing network.Discussion
In this study, two parallel deep-learning networks were trained in a self-supervised manner to synthesize HR images from LR patient images. The reported approach integrates a self-supervised strategy into a deep learning-based HR MR imaging framework without any paired LR-HR training data. This deep learning strategy enables the network to learn and perform the LR-to-HR mapping specifically from the scanner-acquired data for an individual patient. As a result, there is no need to rely on population-based training datasets that may not account for the patient-specific and individually different anatomy.Conclusion
We have shown a patient-specific deep learning-based method with self-supervised parallel networks to synthesize HR MR images without the need for traditional large training datasets or ground truth images. This approach demonstrated that HR image features in the image with LR in the slice direction can be learned from HR images in the two orthogonal directions. The results support the potential of using artificial intelligence to improve image resolution beyond the limits of current MRI data acquisition. The self-supervision capability could have broad clinical applications, especially with regard to shortening MR acquisition time in clinical PET/MRI practices, to enable accelerated MRI without sacrificing spatial resolution.Acknowledgements
No acknowledgement found.References
1. Plenge E, Poot DHJ, Bernsen M, et al. Super-resolution methods in MRI: Can they improve the trade-off between resolution, signal-to-noise ratio, and acquisition time? Magnetic Resonance in Medicine. 2012;68(6):1983-1993.
2. Paganelli C, Whelan B, Peroni M, et al. MRI-guidance for motion management in external beam radiotherapy: current status and future challenges. Physics in Medicine & Biology. 2018;63(22):22TR03.
Figures
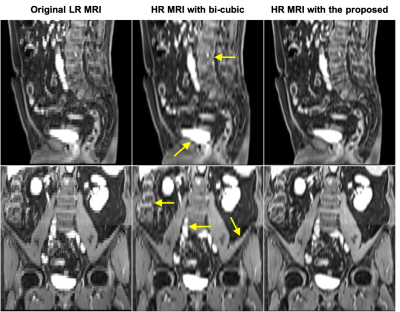
Figure 1. Comparison of original LR images (0.98 mm × 0.98 mm × 6.54 mm) in the cranial-caudal direction (left) and enhanced HR images (0.98 mm × 0.98 mm × 0.98 mm) with bi-cubic interpolation (middle) and with proposed method (right). Images of the sagittal and coronal planes are shown in the first to second rows, respectively.
DOI: https://doi.org/10.58530/2023/4205