4199
Coil2Coil 3D: Improved self-supervised MR image denoising using phased-array coil images based on 3D structure information1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2School of Electrical Engineering, Korea University, Seoul, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
Introduction
MR images suffer from signal-to-noise ratio (SNR) deficiency, hampering accurate diagnosis or quantitative mapping. Recently, deep learning-based denoising methods have been investigated and shown great performance1-2. However, obtaining clean images is difficult or impossible and, therefore, self-supervised learning methods have been proposed3-5. Among them, Coil2Coil5 (C2C) applies Noise2Noise6 (N2N) algorithm, by generating a pair of noisy images from phased-array coil images, showing promising performance, without bounding any specific sequence (i.e., DWI or dynamic MRI). However, C2C was designed with a 2D network, which may not be an optimal structure for 3D MR images, such as multi-echo GRE. In this study, we proposed a 3D denoising network structure based on C2C to improve performance by utilizing 3D structural information.Methods
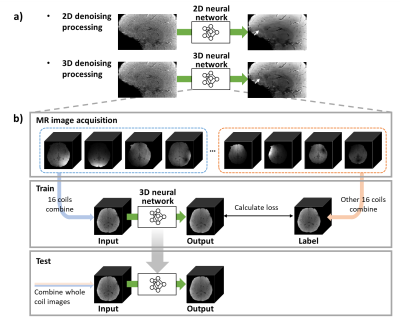
[Coil2Coil 3D]In C2C, 2D phased-array coil images were divided into two different sets, and then, each set was combined two images. After these images were processed to satisfy conditions to apply N2N algorithm, one image was used as input, and the other images used as label. After the training, this method uses the multi-channel combined images as input, generating the denoised combined images as output. By extending the C2C algorithm, a new neural network structure for a 3D MR image denoising was designed by utilizing NAFnet7 as the network structure.
[Dataset]
To train and test the proposed method, complex-valued multi-echo GRE images were acquired (IRB approved) from four subjects (3 subjects for training, 1 subject for test), with 32 phased-array head coils. These images of each subject consisted of 6 head orientations with the following protocols: resolution = 1 × 1 × 1 mm3, TR = 38 ms, TE = 7.70:5.03:32.85 ms (6 echoes). For data preprocessing, each subject was normalized to have a mean of 0 and standard deviation of 1. By using the images from each echoes and directions, a total number of 108 3D images and 36 3D images were used for the training set and test dataset, respectively. The sensitivity maps were estimated using ESPIRiT8.
[Denoising of synthetic noise-added images]
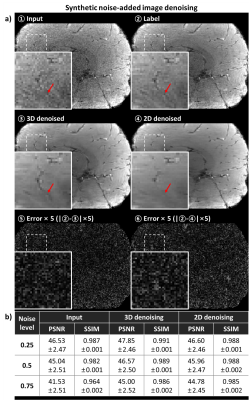
For comparison, the acquired MR images corrupted by adding synthetic noise were used for training and test. The network was trained with a noise level (i.e., a standard deviation of zero-mean Gaussian) of 0.5, and tested with three noise levels of 0.25, 0.5, and 0.75. After the training, the denoised output from the synthetically corrupted images were compared to the coil original images before the corruption. For the quantitative metric, peak signal-to-noise ratio (PSNR) and structural similarity index (SSIM) were calculated.
[Denoising of real-world images]
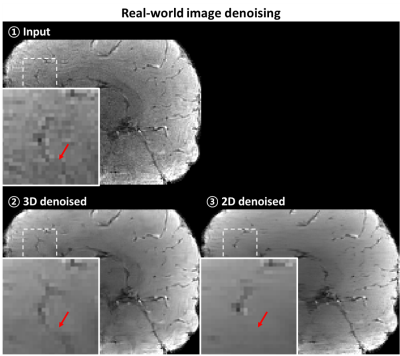
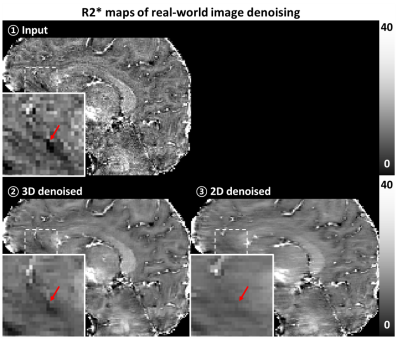
To validate the proposed method for the real-world images, the training and test process were newly conducted for the real-world images. Different to the synthetic noise-added experiment, MR images without any additional noise (i.e., existing only real-world noise) were utilized to either training and test. Since obtaining clear image is unable, qualitative comparison was performed by observing that the detail structures were conserved.
[Quantitative mapping using the denoised images]
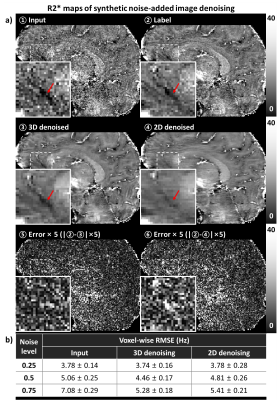
To demonstrate application of the proposed method in quantitative mapping, the denoised multi-echo GRE images were used for R2* mapping. The denoising results of the synthetic noise-added experiment were utilized for quantitative evaluation, and also, the real-world images experiment was performed. The quantification results using denoised images were compared to those of using the images before the denoising. In quantitative comparison, the root-mean-square-error (RMSE) was calculated. For the R2* mapping, a non-linear fitting on mono exponential function was conducted to each voxel from the multi-echo GRE images after each echo images were processed separately.
Results
For synthetic noise-added experiment (Fig. 2), the proposed method using 3D-based network demonstrated successful denoising, showing the improved performance when comparing to the 2D-based network method in all noise levels (case of noise level = 0.5; PSNR: 46.57 ± 2.50 for 3D vs. 45.96 ± 2.47 for 2D, 45.04 ± 2.51 for before denoising, SSIM: 0.989 ± 0.001 for 3D, 0.988 ± 0.002 for 2D, 0.982 ± 2.51 for before denoising). In both case of the synthetic noise and real-world noise (Fig. 4), the qualitatively successful denoising were shown by preserving more structural details while the 2D network does not. In R2* mapping experiment (Fig. 3, 5), the quantitative mapping results using the denoised images showed the reduced quantitative errors than those of images without denoising in case of synthetic noise (case of noise level = 0.5; RMSE: 4.46 ± 0.17 Hz for 3D, 4.81 ± 0.26 Hz for 2D, 5.06 ± 0.25 Hz for before denoising) and qualitative improvement in both case of synthetic noise and real-world noise.Discussion and Conclusion
In this work, a deep-learning-based method using 3D denoising network was proposed. By utilizing the 3D image structure information, the proposed method successfully denoised the MR images while conserving the more tissue details than 2D-based network. This method may be valuable for the diagnosis and quantification by the improved denoising results. The point of that the increased dimension indicated the improved results may be more enhancing the performance in case of more information such as a consideration of echo dimension for multi-echo data and time series for dynamic imaging.Acknowledgements
This work has been supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021M3E5D2A01024795) and Samsung Electronics Co., Ltd.References
[1] L. Gondara, "Medical image denoising using convolutional denoising autoencoders," in 2016 IEEE 16th international conference on data mining workshops (ICDMW), 2016: IEEE, pp. 241-246.
[2] M. Kidoh et al., "Deep learning based noise reduction for brain MR imaging: tests on phantoms and healthy volunteers," Magnetic Resonance in Medical Sciences, vol. 19, no. 3, p. 195, 2020.
[3] S. Fadnavis, J. Batson, and E. Garyfallidis, "Patch2Self: denoising diffusion MRI with self-supervised learning," arXiv preprint arXiv:2011.01355, 2020.
[4] J. Xu and E. Adalsteinsson, "Deformed2Self: Self-Supervised Denoising for Dynamic Medical Imaging," arXiv preprint arXiv:2106.12175, 2021.
[5] J. Park et al., "Coil2Coil: Self-supervised MR image denoising using phased-array coil images," arXiv preprint arXiv:2208.07552, 2022.
[6] J. Lehtinen et al., "Noise2noise: Learning image restoration without clean data," arXiv preprint arXiv:1803.04189, 2018.
[7] L. Chen et al., "Simple baselines for image restoration," arXiv preprint arXiv:2204.04676, 2022.
[8] M. Uecker et al., "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA," Magnetic resonance in medicine, vol. 71, no. 3, pp. 990-1001, 2014.
Figures