4188
Denoising highly accelerated T2*-weighted brain MRI using a deep learning convolutional neural network
Bryan Quah1, Sreekanth Madhusoodhanan Nair1, Jin Jin2, Fei Han3, Brian Renner1, Elaina Gombos1, Ke Cheng Liu3, Sunil Patil3, John A. Derbyshire4, Ken Sakaie5, Emmanuel Obusez5, Jonathan Lee5, Mark Elliot6, Russell T. Shinohara7, Matthew K. Schindler8, Jae W. Song6, Michel Bilello6, Marwa Kaisey1, Nader Binesh9, Marcel Maya9, Javier Galvan9, Hui Han10, Debiao Li10, Andrew Solomon11, Daniel S. Reich12, Nancy L. Sicotte1, Mark Lowe5, Daniel Ontaneda13, Omar Al-Louzi1, and Pascal Sati1,10
1Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Siemens Healthcare Pty Ltd, Brisbane, Australia, 3Siemens Medical Solutions, PA, United States, 4Functional MRI Facility, National Institute of Mental Health, National Institutes of Health, Bethesda, MD, United States, 5Imaging Institute, Cleveland Clinic, Cleveland, OH, United States, 6Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States, 7Department of Biostatistics, Epidemiology, and Informatics, Penn Statistics in Imaging and Visualization Center, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, United States, 8Department of Neurology, University of Pennsylvania, Philadelphia, PA, United States, 9Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 10Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 11Larner College of Medicine, The University of Vermont, Burlington, VT, United States, 12Translational Neuroradiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 13Mellen Center, Department of Neurology, Neurological Institute, Cleveland Clinic, Cleveland, OH, United States
1Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Siemens Healthcare Pty Ltd, Brisbane, Australia, 3Siemens Medical Solutions, PA, United States, 4Functional MRI Facility, National Institute of Mental Health, National Institutes of Health, Bethesda, MD, United States, 5Imaging Institute, Cleveland Clinic, Cleveland, OH, United States, 6Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States, 7Department of Biostatistics, Epidemiology, and Informatics, Penn Statistics in Imaging and Visualization Center, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, United States, 8Department of Neurology, University of Pennsylvania, Philadelphia, PA, United States, 9Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 10Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 11Larner College of Medicine, The University of Vermont, Burlington, VT, United States, 12Translational Neuroradiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 13Mellen Center, Department of Neurology, Neurological Institute, Cleveland Clinic, Cleveland, OH, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Data Processing, Denoising, Neuroimaging
The scan time of high-resolution T2*-weighted brain imaging using 3D echo-planar imaging (3D-EPI) can be significantly reduced by applying Controlled Aliasing In Parallel Imaging Results In Higher Acceleration (CAIPIRINHA). However, this comes at the expense of a significant reduction in image quality. In this study, we evaluated the feasibility of using a deep learning-based approach (DnCNN) to denoise highly accelerated 3D-EPI scans acquired at 3T. Our results show that DnCNN was able to efficiently denoise highly accelerated T2*-weighted brain scans while preserving anatomical and pathological details.Introduction
High-resolution susceptibility-based imaging using 3D echo-planar imaging (3D-EPI) enables the detection of new diagnostic biomarkers of neurological disorders, such as the central vein sign (CVS) and paramagnetic rim lesions (PRL) in multiple sclerosis (MS).1,2,3,4 The recommended 3D-EPI acquisition for optimal image quality at 3T is ~6 minutes long,5 which is suboptimal for a widespread implementation in hospitals and private imaging centers. One strategy to further reduce the scan time is to include advanced parallel imaging techniques, such as Controlled Aliasing In Parallel Imaging Results In Higher Acceleration (CAIPIRINHA).6 However, this comes at the expense of a significant reduction in signal-to-noise ratio (SNR), which typically impacts clinical interpretation. In this study, we evaluated the use of a deep learning-based approach to denoise highly accelerated T2*-weighted 3D-EPI scans acquired at 3T.Methods
3T brain MRI scans from 52 adults scanned at three imaging sites (Cedars-Sinai Medical Center, Cleveland Clinic, University of Pennsylvania) as part of the CAVS-MS study5 were used for the training and testing of a deep learning denoiser. High-resolution (650 mm isotropic) T2*-weighted 3D-EPI (T2* 3D-EPI) scans were acquired with a research sequence without parallel imaging (acquisition time, TA: ~6 minutes) and with parallel imaging using 2D CAIPIRINHA undersampling at different total acceleration factors: R=2 (TA: ~4 minutes), R=3 (TA: ~3 minutes) and R=4 (TA: ~2 minutes).To build the training set, T2* 3D-EPI scans w/o CAIPIRINHA were preprocessed as follows: (1) intensity scaling, (2) removing slices without brain tissue, (3) zero padding, (4) randomly flipping the 3D image horizontally and/or vertically, (5) adding Gaussian noise with standard deviation chosen between 3-5% of the maximum image intensity, and (6) extracting nine 75% overlapping 2D patches of dimension 256 x 256 (Figure 1). The testing set used for evaluating the model prediction was created by repeating steps (1), (3) and (6) on the T2* 3D-EPI scans w/ CAIPIRINHA (Figure 1).
A 2-dimensional denoising convolutional neural network (DnCNN) was used for denoising.7,8 This architecture allowed for end-to-end training and contained 17 layers including convolution, batch normalization, and rectified linear unit operations to denoise any given image (Figure 2). During model prediction, a patch-wise denoising followed by a reconstruction of the full image was performed by stitching the patches together and taking the average for the overlapping regions. The network was trained for 40 epochs with a patience of 5 using the Adam optimizer with mean squared error as the loss function and was tested using a 10-fold cross validation. Network training and prediction was done on an Ubuntu machine with 32-core 3.7 GHz AMD CPU, 256GB memory, and four NVIDIA Quadro RTX 6000 with 24GB video memory.
Peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM) were measured9,10 for each testing image before and after denoising. Additionally, these measures were stratified according to the CAIPIRINHA total acceleration factor to determine how the denoising algorithm performed for each acceleration.
Results
Representative images from a high-resolution T2* 3D-EPI scan acquired with and without CAIPIRINHA at 3T are shown before and after denoising (Figure 3). Denoised images display higher image quality while preserving the visibility of brain features and disease-related biomarkers (PRL and CVS; magnified view in Figure 3). The average PSNR values measured across the entire cohort were substantially increased after denoising for all three acceleration factors (24% increase for R=2; 32% increase for R=3; 41% increase R=4) (Figure 4). Meanwhile, the SSIM values measured across the entire cohort remained high for all three acceleration factors (mean ± SD: 0.986 ± 0.0032 for R=2; 0.984 ± 0.0034 for R=3; 0.982 ± 0.0036 for R=4). For network training, each fold was trained for an average of 18 epochs at an average of ~8 minutes per epoch. On the other hand, model prediction took only ~50 seconds to denoise a single subject and ~2 minutes to denoise a batch of six cases at maximum video memory usage.Discussion
As demonstrated by our initial results, DnCNN was able to efficiently denoise highly accelerated T2*-weighted brain scans while preserving anatomical and pathological details. More specifically, DnCNN was able to compensate for the reduction in SNR occurring at higher R factors by providing increased PSNR while maintaining SSIM close to 1. Interestingly, the higher percentage increase in PSNR obtained at higher acceleration factors indicated the ability of our methodology to remove varying levels of noise present within the images. Another important aspect of our work was to create a training set using artificially noised T2* 3D-EPI scans w/o CAIPIRINHA, which allowed us to independently test the performance of the DnCNN on the accelerated undersampled T2* 3D-EPI scans w/ CAIPIRINHA. Similarly, cross-validation prevented the DnCNN from seeing the same subjects used for training within the testing dataset. This should allow for robust denoising when deploying this methodology on a larger scale.Conclusion
We developed a deep learning approach for improving the image quality of ultra-fast, high-resolution, T2*-weighted imaging at 3T. In this pilot study, we demonstrated the feasibility of our approach in a multicenter dataset. Further studies will be aimed at evaluating our approach for the detection of diagnostic biomarkers of MS in a clinical setting.Acknowledgements
This work was supported by the National Institute of Neurological Disorder and Stroke (NINDS) 1U01NS116776-01 and the National Multiple Sclerosis Society (NMSS) RG-2110-38526.References
- Sati P, Oh J, et al. The central vein sign and its clinical evaluation for the diagnosis of multiple sclerosis: a consensus statement from the North American Imaging in Multiple Sclerosis Cooperative. Nature Reviews Neurology, vol. 12, no. 12, Dec 2016.
- Maggi P, Sati P, et al. Paramagnetic Rim Lesions are Specific to Multiple Sclerosis: An International Multicenter 3T MRI Study. Annals of Neurology, vol. 88, no. 5, pg. 1034-1042, Nov 2020.
- M. Absinta, P. Sati, A. Fechner, M.K. Schindler, G. Nair and D.S. Reich. Identification of Chronic Active Multiple Sclerosis Lesions on 3T MRI. American Journal of Neuroradiology, vol. 39, no. 7. July 2018.
- Kolb H, Al-Louzi O, et al. From pathology to MRI and back: Clinically relevant biomarkers of multiple sclerosis lesions. Neuroimage: Clinical, vol. 36 2022.
- Ontaneda D, Sati P, et al. Central vein sign: A diagnostic biomarker in multiple sclerosis (CAVS-MS) study protocol for a prospective multicenter trial. Neuroimage Clinical, vol. 32 2021.
- Breuer F, Blaimer M, et al. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magnetic Resonance in Medicine. vol 55, no. 3, March 2006.
- Zhang K, Zuo W, Chen Y, Meng Deyu, Zhang L. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Transactions on Image Processing, vol. 26, no. 7, 2017.
- Kidoh M, Shinoda K, et al. Deep Learning Based Noise Reduction for Brain MR Imaging: Tests on Phantoms and Healthy Volunteers. Magnetic Resonance in Medical Sciences, vol. 19, no. 3, 2020.
- Wang Z, Bovik A, Sheikh H, Simoncelli E. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Transactions On Image Processing, vol. 13, no. 4, April 2004.
- Hore A, Ziou D. Image Quality Metrics: PSNR vs. SSIM. International Conference on Pattern Recognition, pp. 2366-2369. August 2010.
Figures
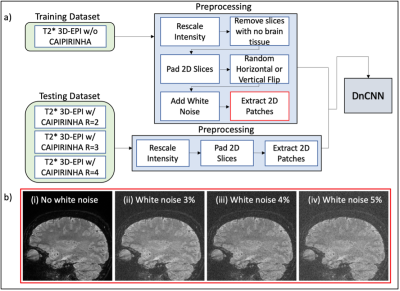
a) T2* 3D-EPI images from the 52 subjects are split into training and testing sets according to their CAIPIRINHA total acceleration factor. Both training and testing datasets underwent similar preprocessing steps before being fed to the convolutional neural network (DnCNN) for training and prediction. b) Examples of patches obtained after the final pre-processing step for training the network: (i) patch before white noise is added; (ii) patch with white noise 3% maximum intensity; (iii) patch with white noise 4% maximum intensity; (iv) patch with white noise 5% maximum intensity.
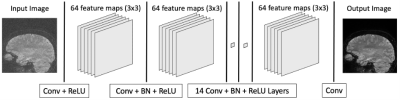
The denoising convolutional neural network (DnCNN) consists of 17 total layers to extract and compose features from the training set. During prediction, the learned weights of each layer contribute to removing noise as the image passes through the network. Conv: Convolution, ReLU: Rectified Linear Unit, BN: Batch Normalization.
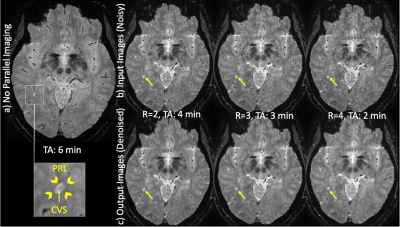
T2* 3D-EPI images from a subject evaluated for a possible diagnosis of MS: a) Original images acquired without CAIPIRINHA; b) Input images acquired with CAIPIRINHA; c) Output images after denoising using DnCNN. PRL: paramagnetic rim lesion, CVS: central vein sign.
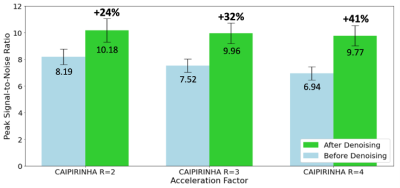
Average peak signal-to-noise ratio with 95% confidence intervals before and after images have been denoised for each CAIPIRINHA total acceleration factor (R=2, 3, 4). The relative increase in PSNR (%) is noted above the plots.
DOI: https://doi.org/10.58530/2023/4188