4164
Feasibility of spiral diffusion imaging on a clinical 3T MR system1BRAIN-TO Lab, Techna Institute, University Health Network, Toronto, ON, Canada, 2UBC MRI Research Centre, Vancouver, BC, Canada, 3Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada, 4Center for Neuroscience Imaging Research, Institute for Basic Science & Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of
Synopsis
Keywords: Diffusion/other diffusion imaging techniques, Diffusion Tensor Imaging, Spiral DWI
Spiral diffusion imaging has been shown to provide substantial SNR advantages (>50%) over state-of-the-art echo-planar imaging, due to the attainable shorter echo times. We investigate the feasibility of this approach on a standard 3T MR system without additional instrumentation. All tools for successful implementation are freely made available, i.e., the characterization measurement for the gradient system using a standard phantom, and an open-source, fast image reconstruction suite in Julia correcting for gradient imperfections, eddy currents, and static B0 off-resonance. We show that this low-cost, accessible solution provides high-resolution diffusion images (1.1mm) in-vivo with reduced geometric distortion and improved quantitative maps.Introduction
Spiral diffusion imaging has been shown to provide substantial SNR advantages (>50%) over state-of-the-art echo-planar imaging (EPI), due to the attainable shorter echo times and better parallel imaging suitability (reduced g-factor penalty) [1]. To achieve high spiral image quality, current solutions often rely on external measurements of spiral and diffusion encoding gradients to correct for eddy currents and trajectory imperfections. This requires additional hardware (e.g., magnetic field probes [2]). Furthermore, while different solutions for image reconstruction with such corrections exist [3,4], implementations are often not open-source software.In this work, we investigate the feasibility of spiral diffusion imaging on a standard 3T MR system without additional instrumentation. All tools for successful implementation are freely made available, in particular a characterization measurement for the gradient impulse response function (GIRF) of the system using a standard phantom, as well as an open-source, fast image reconstruction suite in Julia correcting for gradient imperfections, eddy currents, and static B0 off-resonance. We evaluate this open-source framework for single-shot high-resolution diffusion imaging (1.1mm) in-vivo and quantify the reduction in geometric distortion and improvement in image quality, as well as the quantitative impact on the calculated diffusion maps.
Methods
SetupTwo healthy volunteers (m, age=35-39y) were scanned on a 3T Siemens Prisma MR system, utilizing the vendor 20-channel receive head-neck coil and body coil excitation.
Sequence, Trajectories and Image Reconstruction
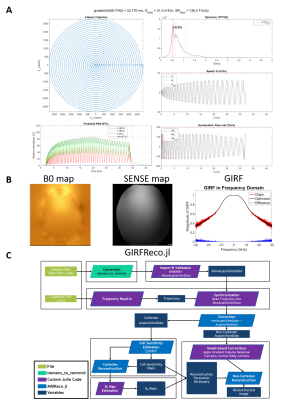
We designed a 2D single shot spiral trajectory (4x undersampled FOV 220 mm) with 1.1mm in-plane resolution and time-optimal readout (32ms, Fig.1A) for our gradient slew-rate and amplitude limits [5], and respecting the SAFE model for peripheral nerve stimulation [6]. In total, 15 transverse slices (2mm thickness, 8mm gap) were acquired covering the brain with TR=3s and TE=40ms.
We acquired two different sets of diffusion-weighted spin-echo spiral images with unipolar diffusion-sensitized gradients: (i) using a b-value of 2000 s/mm2 with 6 diffusion directions and a b=0 image (4 averages, total scan time: 1:24 min), and (ii), b=1000 s/mm2 with 30 directions and a b=0 image (4 averages, total scan time: 6:12 min).
A spin-warp dual-echo sequence (TE1/2 4.93/7.38 ms, TR 0.4s, flip angle 60 deg) was measured prior to the diffusion scans with identical geometry to serve as anatomical reference, as well as to estimate sensitivity and off-resonance map for the expanded signal model (Fig. 1B, [4]) using ESPIRIT [7] and regularized field map estimation [8], respectively.
The MR signal model for image reconstruction comprised these maps and the spiral field dynamics (k0, kx,y,z), as predicted from the nominal spiral gradient waveform via a gradient impulse response function (GIRF, [9]). The GIRF was previously characterized for this system using a phantom-based measurement [10]. Data processing and image reconstruction were performed using the Julia package GIRFReco.jl developed in-house (Fig.1, [11], download of all reconstruction code https://github.com/brain-to/GIRFReco.jl), which relies on the MRIReco.jl package [12] for the core iterative cg-SENSE reconstruction algorithm with time-segmented static off-resonance correction [13,14].
Diffusion Analysis
Fractional anisotropy (FA), mean diffusivity (MD) and the diffusion tensor eigenvector/value images (EV1-3) maps were calculated using FSL dtifit following the MP-PCA denoising using MRTrix3 [15] dwidenoise. No eddy preprocessing was done since it only applies to EPI encoding.
Results
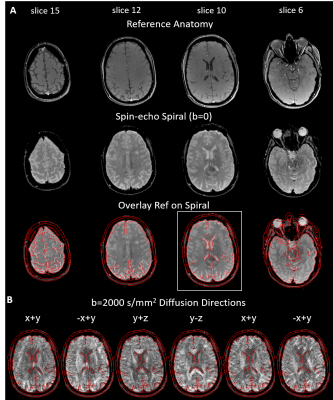
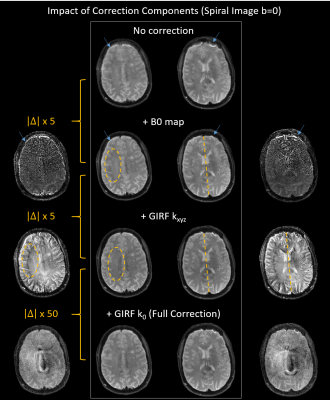
Fig.2 demonstrates that the provided framework delivers overall good spiral image quality for the b=0 image (2A). Few artifacts are visible (mostly in lower slices), and high geometric congruency to the anatomical reference, outlined by its overlaid contour edges. Even for the high-diffusion weighting case (i) (b=2000 s/mm2), spiral image quality remains high and geometric distortions due to diffusion eddy currents appear to be small (2B).Fig.3 scrutinizes the impact of the different correction components. Without corrections (nominal trajectory, no B0 map), even the b=0 image appears to be blurry with concentric spiral PSF off-resonance artifacts (blue arrows). The B0 map-based offresonance correction removes most of these artifacts (consecutive absolute difference images to previous correction left/right, max 20%), with a geometric mismatch (rotation) to the reference image remaining (see yellow dashed line). Implementing a GIRF correction for the trajectory kxyz rectifies this discrepancy (pointing to a gradient delay correction) and eliminates a subtle reconstruction artifact (yellow dashed ellipse and difference images). Correction for the k0 eddy with the GIRF resulted in a small improvement of the central artifact resembling spiral undersampling.
Fig.4 evaluates how spiral image quality propagates into the quantitative diffusion maps. MD, FA and EV1 image show clear high-resolution features, e.g., in white matter tracts of the corpus callosum or surrounding the thalamus.
Discussion
High-resolution, single-shot diffusion imaging (1.1mm) was accomplished with high image quality on a standard clinical 3T MR system, without the need for additional hardware, and implemented as a fast open-source reconstruction framework in Julia. We did not correct for higher order diffusion eddy currents here, which has been shown to further improve geometric accuracy of spiral diffusion images [16]. In lieu of additional instrumentation, this requires a higher-order phantom-based measurement of the eddy currents or the GIRF itself [17]. Further work will compare the spiral diffusion images to state-of-the-art product EPI diffusion sequences and quantify the net SNR benefit for clinical practice.Acknowledgements
No acknowledgement found.References
[1] Y. Lee et al., Magnetic Resonance in Medicine, 2021, 85, 1924.
[2] N. De Zanche et al., Magnetic Resonance in Medicine, 2008, 60, 176―186.
[3] R.K. Robison et al., Magnetic Resonance in Medicine, 2019, 0.
[4] B.J. Wilm et al., Magnetic Resonance in Medicine, 2011, 65, 1690.
[5] M. Lustig, S.-J. Kim, J.M. Pauly, IEEE Transactions on Medical Imaging, 2008, 27, 866.
[6] F. Szczepankiewicz, C.-F. Westin, M. Nilsson, Journal of Neuroscience Methods, 2021, 348, 109007.
[7] M. Uecker et al., Magnetic Resonance in Medicine, 2013, n/a.
[8] A.K. Funai et al., IEEE transactions on medical imaging, 2008, 27, 1484.
[9] S.J. Vannesjo et al., Magnetic Resonance in Medicine, 2013, 69, 583.
[10] Z. Wu et al., in Proc. Intl. Soc. Mag. Reson. Med. 30, 0641.
[11] A. Jaffray et al., in Proc. Intl. Soc. Mag. Reson. Med. 30, 2022, 2435.
[12] T. Knopp, M. Grosser, Magnetic Resonance in Medicine, 2021, 86, 1633.
[13] K.P. Pruessmann et al., Magnetic Resonance in Medicine, 2001, 46, 638―651.
[14] T. Knopp et al., Medical Imaging, IEEE Transactions on, 2009, 28, 394.
[15] J.-D. Tournier et al., NeuroImage, 2019, 202, 116137.
[16] B.J. Wilm et al., Magnetic Resonance in Medicine, 2017, 77, 83.
[17] J. Rahmer et al., Magnetic Resonance in Medicine, 2019, 82, 2146.
Figures