4116
Deep Learning-accelerated, Single-breath-hold T2-weighted Imaging for Tumor Invasion Assessment in Gastric Cancer
Wei-Yue Xu1, Qiong Li1, Ya-Jun Hou1, Yi-Cheng Hsu2, Dominik Nickel3, Yu-Dong Zhang1, and Xi-Sheng Liu1
1Department of Radiology, the First Affiliated Hospital with Nanjing Medical University, Nanjing, China, 2MR Collaboration, Siemens Healthineers Ltd, Shanghai, China, 3MR Applications Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany
1Department of Radiology, the First Affiliated Hospital with Nanjing Medical University, Nanjing, China, 2MR Collaboration, Siemens Healthineers Ltd, Shanghai, China, 3MR Applications Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany
Synopsis
Keywords: Contrast Mechanisms, Contrast Mechanisms, deep learning-accelerated T2WI
T2-weighted imaging (T2WI) is an indispensable sequence of gastric magnetic resonance imaging (MRI) for tumor assessment. This study compared the deep learning-accelerated T2WI sequence (DL-T2WI) with BLADE T2WI in image quality assessment and tumor invasion evaluation of gastric cancer (GC). It revealed that DL-T2WI, acquired within a single-breath-hold, performed better than BLADE T2WI in terms of image quality for both quantitative and qualitative analyses. Furthermore, DL-T2WI images displayed comparable accuracy to BLADE T2WI images for serous invasion evaluation. The study suggests that DL-T2WI might be superior to BLADE T2WI for GC.Introduction and aim
Magnetic resonance imaging (MRI) plays an increasingly vital role in gastric cancer (GC) with the continued technological improvements in abdominal imaging 1. A previous study demonstrated that T2-weighted imaging (T2WI) was of great interest when considering mucinous carcinoma and serous invasion 2. However, peristalsis-related artifacts and motion artifacts remain technical challenges for T2WI sequences in patients with GC. Therefore, rapid scanning with high-quality images is a major area of interest in gastric MRI. In general, respiratory-triggered T2WI, such as BLADE T2WI, is used to maintain image quality at the cost of a longer acquisition time. Recently, a new technique was proposed to rapidly acquire T2WI: deep learning-accelerated single-breath-hold T2-weighted imaging (DL-T2WI) 3. This study aimed to (a) explore DL-T2WI as a potential substitute for BLADE T2WI in gastric imaging and (b) investigate the use of DL-T2WI for identifying the serous invasion status of GC.Method
From August 2022 to October 2022, 25 patients with biopsy-proven GC were prospectively enrolled, who underwent MRI scanning, including DL-T2WI and BLADE T2WI before surgery on a 3T system (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany). The BLADE T2WI parameters were as follows: TR/TE = 4100/98 milliseconds; FOV = 340 × 340 mm2; voxel size = 1.1 × 1.1 × 3.0 mm3; 60 slices; flip angle = 116o; and acquisition time = 4–6 minutes. We used the research application, deep learning-accelerated HASTE 2 for DL-T2WI, and the parameters were as follows: TR/TE = 390/96 milliseconds; FOV =360 × 281 mm2; voxel size = 1.1 × 1.1 × 3.0 mm3; 45 slices; flip angle=116o; and acquisition time = 18 seconds. A combination of quantitative and qualitative approaches was used to assess the image quality for DL-T2WI and BLADE T2WI by 2 experienced radiologists. Figure 1 depicts an image example for quantitative analysis, including signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR). The qualitative analysis included lesion delineation and total image quality (on a Likert scale of 1–5, 1 = very poor, 2 = poor, 3 = moderate, 4 = good, and 5 = excellent). Furthermore, 2 radiologists evaluated the serous invasion status of GC using DL-T2WI and BLADE T2WI (Fig. 2). The intraclass correlation coefficient (ICC) and kappa statistic were used to evaluate the interobserver agreement. The image quality was compared between the 2 methods using the chi-square test or Wilcoxon signed-rank test. The diagnostic accuracy of 2 sequences in identifying the serous invasion status of GC was compared using the chi-squared test.Results
Figure 2 shows sharper boundaries between different tissues and better lesion delineation using DL-T2WI. The ICC and kappa values indicated good interobserver agreement for all parameters (>0.75). For image quality analysis, the SNR and CNR of the DL-T2WI sequence were significantly higher than those of the BLADE T2WI sequence (all P < 0.001). The lesion delineation of DL-T2WI was also significantly better than that of BLADE T2WI (P < 0.01) (Fig. 3). The diagnostic accuracy of BLADE T2WI and DL-T2WI for the serous invasion was 70% and 75%, respectively, with no significant difference (P = 0.795).Discussion
The issue of fast acquisition time and high-quality images in gastric T2WI imaging has received considerable attention. Current data highlighted that DL-T2WI considerably outperformed the BLADE-T2WI in terms of image quality and acquisition time. Also, accurate preoperative prediction of serous invasion in patients with biopsy-proven GC is critical for clinical decision-making. The results of this study indicated that BLADE T2WI and DL-T2WI could identify the serous invasion status with equally excellent performance.Conclusion
DL-T2WI exhibits promising performance compared with BLADE T2WI in terms of image quality and acquisition time.Acknowledgements
No acknowledgement found.References
1. Zhang Y, Yu J. The role of MRI in the diagnosis and treatment of gastric cancer. Diagn Interv Radiol 2020; 26(3): 176-82.
2. Borggreve AS, Goense L, Brenkman HJF, et al. Imaging strategies in the management of gastric cancer: current role and future potential of MRI. Br J Radiol 2019; 92(1097): 20181044.
3. Herrmann J, Gassenmaier S, Nickel D, et al. Diagnostic Confidence and Feasibility of a Deep Learning Accelerated HASTE Sequence of the Abdomen in a Single Breath-Hold. Invest Radiol 2021; 56(5): 313-9.
Figures
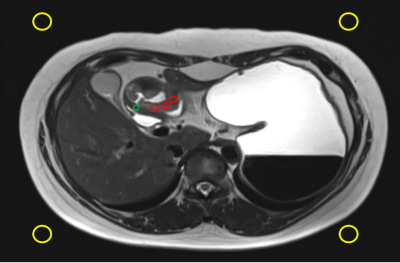
Figure 1. Schematic diagram of ROI. SI_tumor = mean signal intensities of 3 small circles (red); SD (the standard deviation of the noise) = mean signal intensities of the 4 small circles (yellow); SI_gastric wall = mean signal intensities of the normal gastric wall (green). SNR=SI_tumor/SI_ gastric wall; CNR=(SI_tumor-SI_ gastric wall)/SD.
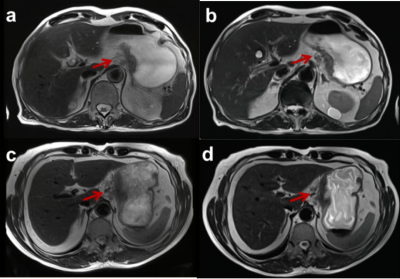
Figure 2. Two patients with gastric cancer with serous invasion who underwent both BLADE T2WI (a and c) and deep learning-accelerated T2-weighted imaging (DL-T2WI) (b and d). Both methods detected a tumor with an irregular serous surface and infiltration in the perigastric plane. The lesion delineation of DL-T2WI (5 scores) was better than that of BLADE T2WI (3 scores).
Figure 3. Image quality assessment using quantitative and qualitative approaches. (a and b) The signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) of the BLADE-T2WI sequence were significantly lower than those of the DL-T2WI sequence (P < 0.001). (c) The lesion delineation score of the DL-T2WI sequence was significantly higher than that of the BLADE-T2WI sequence (P < 0.01).
DOI: https://doi.org/10.58530/2023/4116