4096
Hyperpolarized 13C Metabolic Imaging of the Human Abdomen with Spatiotemporal Denoising1Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2UC Berkeley-UCSF Bioengineering Program, University of California San Francisco, San Francisco, CA, United States
Synopsis
Keywords: Data Processing, Hyperpolarized MR (Non-Gas), Pancreas, Abdomen, Cancer
A substantial challenge in hyperpolarized (HP) 13C MRI is the limited signal-to-noise ratio (SNR) of downstream metabolites, which restricts the achievable spatial resolution. To overcome this for large coverage abdominal studies, a patch-based spatiotemporal denoising approach was applied to denoise dynamic imaging data in [1-13C]pyruvate echo-planar imaging (EPI) human datasets. With denoising, a 11.4 ± 1.8 and 8.7 ± 2.4 fold sensitivity gain was achieved for [1-13C]alanine and [1-13C]lactate, along with improved spatial coverage. These results support the potential of spatiotemporal denoising to improve quantification in HP 13C MRI for normal and cancer studies.Introduction
Hyperpolarized (HP) 13C MRI is a rapid and noninvasive imaging technique that visualizes dynamic metabolic processes and can be used to detect disease and treatment response by measuring the real-time metabolic conversion of HP substrates to downstream metabolites [1]. Despite dynamic nuclear polarization producing >10,000-fold MR signal enhancement, the spatial resolution of HP MRI is limited by the signal-to-noise ratio (SNR) for metabolic products such as [1-13C]alanine and [13C]bicarbonate in HP [1-13C]pyruvate studies. Background noise in the HP spectra can also lead to reduced accuracy of the quantification metabolic biomarkers, such as the first-order enzymatic conversion rates of pyruvate to lactate (kPL) and pyruvate to alanine (kPA). Coil sensitivity and variable rates of perfusion along with low metabolite SNR pose major challenges for HP abdominal imaging applications—including pancreatic ductal adenocarcinoma (PDA), hepatocellular carcinoma (HCC), renal cell carcinoma (RCC), and body metastatic cancers [2].Prior studies explored the benefits of patch based higher-order singular value decomposition (GL-HOSVD) spatiotemporal denoising [3, 4] to improve image quality and quantification in dynamic HP-13C MR studies in the human brain [5, 6]. Here, this project was designed to investigate methods to characterize the spatiotemporal GL-HOSVD technique for improved denoising of HP-13C echo-planar imaging (EPI) data specialized for imaging the human abdomen (healthy volunteers and patients with PDA) acquired using HP [1-13C]pyruvate and a multi-channel array.
Methods
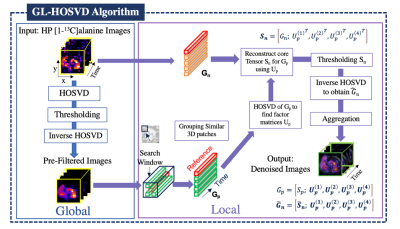
The spatiotemporal patch-based GL-HOSVD denoising technique exploits spatial correlations throughout the image to further improve SNR [3,4]. With this approach, similar 3D patches are grouped into stacks and are processed using hard thresholding to extract a noise-suppressed core tensor. This patch-wise denoising is then carried out across the image domain by moving the search window through the input dynamic data for each metabolite. The final denoised images are then reconstructed by aggregating the multiple estimates at each pixel (Fig. 1). The code that outlines this approach and supports these findings is available at https://github.com/UCSF-HMTRC/hp13c_EPI-hosvd_denoising. The tradeoff between denoising and image fidelity are primarily affected by the two hyperparameters kglobal and klocal. The image quality and accuracy of quantitative analyses following denoising were first evaluated using the simulated [1-13C]pyruvate and [1-13C]lactate dynamics at different noise levels to determine optimal hyperparameters.The spatiotemporal denoising method with the optimized parameters was then applied to two HP [1-13C]pyruvate multi-slice EPI abdominal human cohorts (n = 7 healthy volunteers and n = 8 PDA patients) acquired with a flexible vest coil for RF transmit and an 8-channel array for receive. The mean total SNR (area under the curve SNR, SNRAUC) improvement was quantified, and voxel-wise kinetic modeling was performed on both non-denoised and denoised data to compare the number of voxels quantifiable based on the original SNRAUC > 10 for pyruvate and original SNRAUC > 5 lactate and alanine, along with fitting (<30% relative error) criteria.
Results
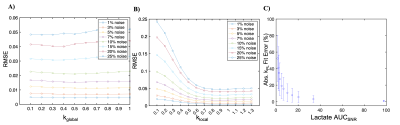
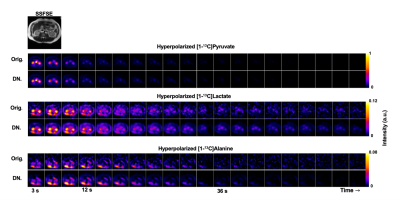
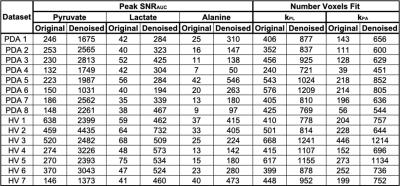
To select adequate hyperparameters for abdominal HP studies, denoising performance was first evaluated using simulated [1-13C]pyruvate and [1-13C]lactate dynamics at different noise levels. Figure 2 indicates that there is a broad range of kglobal : 0.2-0.4 and klocal : 0.6-0.9 values which can denoise HP data while retaining image fidelity when evaluated by RMSE. At the parameterization of kglobal = 0.2 and klocal = 0.9, the kPL bias is <15% when there is adequate SNR (SNRAUC > 5) for downstream metabolites lactate and alanine (Fig. 2 C). Example denoising results for the HP [1-13C]pyruvate EPI data acquired from a healthy volunteer (Fig. 3), and the effective noise reduction seen in [1-13C]alanine dynamics in a pancreatic cancer patient (Fig. 4) show the improvement in image quality after patch-based denoising. The average computation time for denoising one EPI dataset (3 metabolites x 9 slices x 20 timepoints, matrix size of 16x16) using a patch size of 5x5 was 13.2 seconds. Table 1 summarizes the denoising results from the 15 abdominal subjects. In both cohorts, there was mean 9.6 ± 3.3, 8.7 ± 2.4 and 11.4 ± 1.8 fold gain in peak pyruvate, lactate, and alanine SNRAUC respectively. These figures indicate a substantial improvement in the dynamic signals, which then enabled robust quantification of enzymatic rate constants kPL and kPA. Denoising resulted in a 2.1 ± 0.4 fold increase in the number of voxels adequately fit for mapping kPL and a 4.8 ± 2.5 fold increase in the number of voxels for mapping kPA.Discussion and Conclusion
This study demonstrated that spatiotemporal denoising greatly improves visualization of low SNR metabolites and quantification of pyruvate metabolism in dynamic HP-13C MRI studies of the human abdomen. This improvement in SNR and image quality supports the acquisition of HP-13C MRI data at higher spatial resolution in future clinical studies, which would help to reduce partial volume effects and better visualize tumor heterogeneity. Finally, the improvement in the number of voxels which can be used in the quantification of enzymatic rate constants may improve the clinical management of patients with abdominal cancers such as PDA, RCC, and HCC.Acknowledgements
This work was supported by NIH grants U01EB026412, P41EB013598, R01DK115987, and a UCSF Resource Allocation Program Grant. We would also like to acknowledge Mary Frost, Kimberly Okamoto, and Evelyn Escobar for their assistance with the patient studies.References
1. Wang ZJ, et al. Hyperpolarized 13C MRI: State of the Art and Future Directions. Radiology 2019; 291:273–284.
2. Gordon JW et al. Hyperpolarized 13C Metabolic Imaging of Patients with Pancreatic Ductal Adenocarcinoma. ISMRM 2022, London. Abstract ID: 0368.
3. De Lathauwer L, De Moor B, Vandewalle J. A Multilinear Singular Value Decomposition. SIAM J. Matrix Anal. & Appl. 2000;21:1253–1278 doi: 10.1137/S0895479896305696.
4. Zhang X, Peng J, Xu M, et al. Denoise diffusion-weighted images using higher-order singular value decomposition. NeuroImage 2017;156:128–145 doi: 10.1016/j.neuroimage.2017.04.017.
5. Kim Y, et al. Denoising of hyperpolarized 13C MR images of the human brain using patch-based higher-order singular value decomposition. Magn Reson Med. 2021; 86: 2497-256.
6. Vaziri S, et al. Assessment of higher-order singular value decomposition denoising methods on dynamic hyperpolarized [1-13C]pyruvate MRI data from patients with glioma. Neuroimage Clin. 2022; 36: 103155. PMCID: PMC9421383
Figures