4078
7T to 3T domain adaptation in white matter lesion segmentation on T2-weighted (T2-w) FLAIR images using deep learning1Bioengineering, University of Pittsburgh, Pittsburgh, PA, United States, 2Electrical and Computer Engineering, University of Pittsburgh, Pittsburgh, PA, United States, 3Temple University, Philadelphia, PA, United States, 4Psychiatry, University of Pittsburgh, Pittsburgh, PA, United States
Synopsis
Keywords: High-Field MRI, Machine Learning/Artificial Intelligence
In this work, we explored the domain adaptation problem in deep learning segmentation. Specifically, we applied the residual U-net [1] on 3T and 7T Fluid Attenuated Inverse Recovery (FLAIR) images to delineate the white matter hyperintensity (WMH) in a 2D fashion. We leveraged learning without forgetting [2] to regulate the network’s learning in the new domain to preserve the model’s performance on the old domain while still achieving satisfying results on the new domain images.Introduction
Many studies have shown success in segmenting white matter lesions leveraging deep learning models on 3T FLAIR images [3, 4]. However, little to no studies have explored the segmentation performance on 7T FLAIR images using pretrained 3T FLAIR models. One important thing to note in leveraging pretrained models is that the model may suffer catastrophic forgetting in that the adjusted weights are no longer suited for the old domain data and the model has “forgotten” what it had learned previously. Several works have provided some remedies to alleviate the catastrophic forgetting in transfer learning such as learning without forgetting[2], and elastic weight consolidation[5].In this work, we explored the domain adaptation in white matter hyperintensity segmentation in two directions, namely from 3T FLAIR to 7T FLAIR and from 7T FLAIR to 3T FLAIR. We implemented the pseudo label loss mentioned in [2] to prevent the model from forgetting what it had learned in the previous domain.
Methods
We used in-house collected 3T and 7T FLAIR images at the University of Pittsburgh. The resolutions for the two domains are 1x1x3mm (3T) and 0.75x0.75x1.5mm (7T) respectively, and the total numbers of axially sliced 2D FLAIR images for each domain are 6888 (3T) and 5292 (7T). The collected FLAIR images are of size 212x256x48 for the 3T domain and 256x256x108 for the 7T domain. The 3T data includes 160 T2-w FLAIR scans (average age 76, 105 females, 55 male), and the 7T data includes 49 T2-w FLAIR scans (average age 68, 35 females, 14 males).The FLAIR images and the corresponding hand labeled WMH masks were then split into training and validation sets with an 80-20 ratio. We implemented the residual U-net proposed in [1] to perform WMH segmentation. In the pre-training phase, both 3T FLAIR and the 7T FLAIR models were trained independently with a batch size of 32, learning rate of 0.0001, Adam optimizer, dice score loss function, and a reduce-on-plateau learning rate scheduler. After the pre-training phase, pseudo labels were generated by feeding the new domain training set images to the pre-trained model. For the domain adaptation, two losses were constructed: 1) the dice score loss between the output and the actual label, and 2) the dice score loss between the output and the pseudo label. By adjusting the importance weighting of two losses, we could regulate the model’s learning on the new domain data.
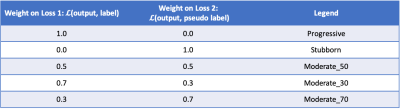
We further performed several experiments using the following importance weights on the two losses (Table 1). An importance weight of 1 on loss2 indicates that the model wants to preserve all the knowledge that it had learned previously, whereas an importance weight of 1 on loss 1 indicates that the model will adjust the pre-trained model’s parameters solely based on the new domain data.
Results
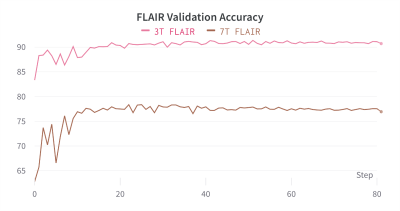
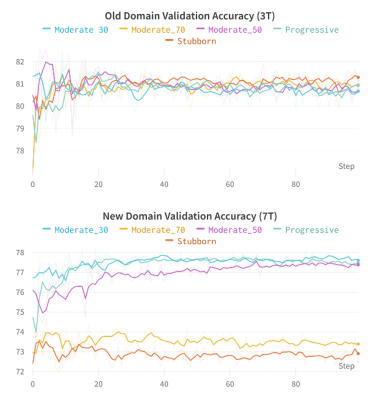
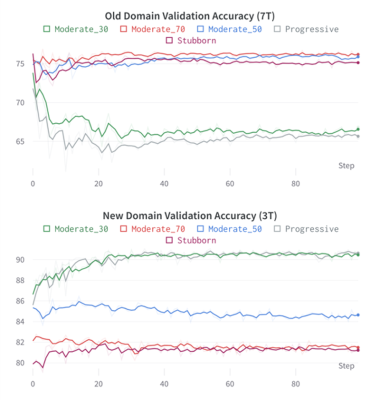
Figure 1 shows the two models segmentation performance on each domain’s validation data. After the pre-training phase, the 7T FLAIR model can reach a validation accuracy of 77%, and the 3T FLAIR model reaches a validation accuracy of 91% (Figure 2). Figure 3, and 4 shows the validation accuracy for different scenarios using the different importance weights listed in the table. When adapting from the pre-trained 7T FLAIR model, an equal split of importance for the two losses might be the best option, as the model have satisfying results in both domains. Interestingly, when adapting from the pre-trained 3T FLAIR model, all scenarios seem to have similar performance on the old domain testing data. Additionally, an equal split of importance for the two losses gave indistinguishable validation performance on the new domain data.Discussion and Conclusions
In this work, we first trained residual U-net models to perform white matter hyperintensity segmentations on 3T and 7T FLAIR images independently. Further, we leveraged the concept presented in the previous work [1] and explored domain adaptation using pre-trained models in both directions, namely 3T FLAIR adapting to 7T FLAIR pre-trained model and 7T FLAIR adapting to 3T FLAIR pre-trained model.Our results demonstrated successful domain adaption on white matter hyperintensity segmentation while leveraging pseudo label loss that preserves the model performance on the old domain yet achieving satisfying results in the new domain.
Acknowledgements
This work was supported by the National Institutes of Health under award number: R01AG067018, RF1AG025516, R01MH111265, R01AG063525, R56AG074467. This work was also supported in part by the University of Pittsburgh Center for Research Computing through the resources provided.References
1. Ding, P.L.K., et al. Deep residual dense U-Net for resolution enhancement in accelerated MRI acquisition. in Medical Imaging 2019: Image Processing. 2019. SPIE.
2. Li, Z. and D. Hoiem, Learning without forgetting. IEEE transactions on pattern analysis and machine intelligence, 2017. 40(12): p. 2935-2947.
3. Khademi, A., et al., Segmentation of white matter lesions in multicentre FLAIR MRI. Neuroimage: Reports, 2021. 1(4): p. 100044.
4. Sundaresan, V., et al., Triplanar ensemble U-Net model for white matter hyperintensities segmentation on MR images. Medical image analysis, 2021. 73: p. 102184.
5. Kirkpatrick, J., et al., Overcoming catastrophic forgetting in neural networks. Proceedings of the national academy of sciences, 2017. 114(13): p. 3521-3526.
Figures