4069
Automatic Segmentation of Rotator Cuff Muscles on MR Images Using Deep Learning1Radiology, University of Washington, Seattle, WA, United States
Synopsis
Keywords: Muscle, MSK, Deep learning
Rotator cuff (RC) injuries are a common occurrence affecting millions of people across the globe. Quantitative MRI-based evaluation of RC injuries can aid in early diagnosis and improve the treatment outcome. A crucial step towards developing a quantitative, clinically relevant methods for these patients, is developing reliable automatic techniques for segmentation of RC muscles. In this study, we developed a deep convolutional neural network model to automatically segment RC muscles on T1-weighted MR images. We showed that the proposed deep learning method provides rapid and reliable automatic segmentation of RC muscles, with an accuracy comparable with that of human raters.
Background or Purpose
The rotator cuff (RC) is an important structural component in the shoulder enabling a wide range of movements and preserving shoulder stability. Subscapularis, infraspinatus, supraspinatus, and teres minor are the muscles that make up the rotator cuff. Rotator cuff injuries are common debilitating disorders with a significant burden. Quantitative analysis of these muscles can improve the accuracy and reliability of the diagnosis and treatment planning. However, the clinical use of quantitative methods has been hampered by the time constrains in busy clinical workflows. Rapid and reliable segmentation of RC muscles is the first step toward the creation of fully automated quantitative diagnostic or prognostic models. Therefore, in this study we developed and validated a residual deep convolutional encoder-decoder U-net network to automatically segment rotator cuff muscles on T1 weighted MR images.Methods
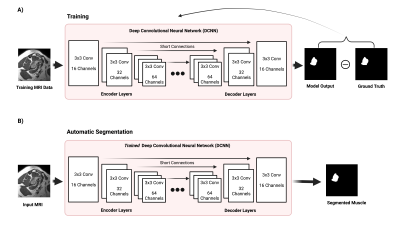
We trained a residual encoder-decoder U-net DCNN model [1,2] depicted in Figure 1. In this architecture, images are decomposed from low-level to high-level step by step to extract essential information from the images. This deep network implicitly learns the anatomical features of muscles from MRI data. This residual encoder-decoder DCNN model includes three encoder/decoder layers. Each encoder layer consists of three 2D convolutional layers with a 3x3 kernel, a batch normalization layer, a rectifier linear unit (ReLU) activation layer, and a 2x2 maximum pooling (down-sampling) layer. A residual connection is placed at the central layer. Each decoder layer consists a 2x2 up-sampling layer followed by three 2D convolutional layers and ReLU activation layers. Bypassing shortcuts connect corresponding encoder-decoder layers to retain high-resolution information in the images.We trained the model using T1 weighted MR images of 124 subjects (training data + validation data) and evaluated the trained model on the other 33 subjects (test data). Both training and test datasets were a mixture of normal and partial RC tear cohorts. During training, the DCNN learns the nonlinear image transformation from input MRI to RC muscles masks and adaptively update its parameters to minimize the a loss function. Binary cross entropy (BCE) [3] and GAN loss functions [4] were used and compared. We employed data augmentation in training images to avoid overfitting. We also utilized the dropout technique to improve the DCNN’s generalization capability. Grid search was performed to find optimum hyperparameters. To quantitatively assess the model performance, we quantified the similarity between the model outputs and ground-truth manual segmentations using the Dice score [5].
Results
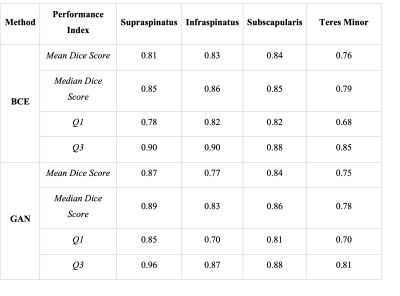
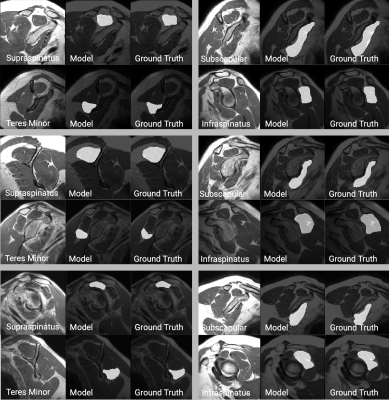
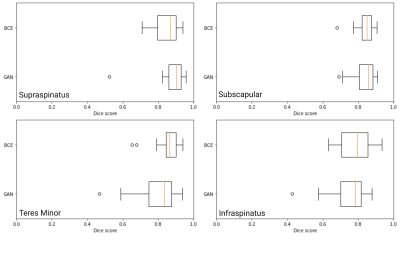
For the supraspinatus muscle, median Dice score was 0.85 (Q1: 0.78, Q3:0.9) for the BCE models and 0.89 (Q1: 0.85, Q3:0.96) for the GAN model. For the subscapularis muscle, the median dice score for the BCE model was 0.85 (Q1: 0.82 , Q3:0.88) compared to a median dice score of 0.86 (Q1: 0.81 , Q3:0.88) on the GAN model. For the infraspinatus muscle, the BCE model achieved a median dice score of 0.86 (Q1: 0.82, Q3:0.90) while the GAN model reached 0.83 (Q1: 0.70, Q3:0.87). Statistics on model performance for all muscles are available in Figure 2. Figures 3 contains examples of the segmented muscles (Subscapularis, infraspinatus, supraspinatus, and teres minor) obtained using the proposed method compared with the ground-truth manual segmentation. Figure 4 contains box plots comparing model performance quantified using the Dice coefficient for each muscle. In this study, Binary Cross Entropy loss function provided improved segmentation accuracy compared with GAN loss function.Conclusions
In this work, we proposed a deep learning method based on residual U-Nets architecture that can accurately segment individual RC muscles (Subscapularis, infraspinatus, supraspinatus, and teres minor) in healthy and partial RC tear patients. To improve the accuracy and generalisablity of the model, during training, we used data from a combination of healthy individuals and patients that were scanned using different MR machines. We also evaluated different loss functions to optimize the performance of the method. This model can be used in the future to create more advanced automated image analysis pipelines for shoulder injuries.Acknowledgements
This work is supported by the Garvey Institute for Brain Health Solutions, and NIH grant 1K01AG071798-01.References
[1] Guo J, Gong E, Fan AP, Goubran M, Khalighi MM, Zaharchuk G. Predicting (15)O-Water PET cerebral blood flow maps from multi-contrast MRI using a deep convolutional neural network with evaluation of training cohort bias. J Cereb Blood Flow Metab. 2019:271678X19888123. Epub 2019/11/15. doi: 10.1177/0271678X19888123. PubMed PMID: 31722599.9.
[2] Chen H, Zhang Y, Kalra MK, Lin F, Chen Y, Liao P, Zhou J, Wang G. Low-Dose CT With a Residual Encoder-Decoder Convolutional Neural Network. IEEE Trans Med Imaging. 2017;36(12):2524-35. Epub 2017/06/18. doi: 10.1109/TMI.2017.2715284. PubMed PMID: 28622671; PMCID: PMC5727581.
[3] Jadon S. A survey of loss functions for semantic segmentation. In: 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) [Internet]. Via del Mar, Chile: IEEE; 2020
[4] J. Yang, D. Ruan, J. Huang, X. Kang and Y. -Q. Shi, "An Embedding Cost Learning Framework Using GAN," in IEEE Transactions on Information Forensics and Security, vol. 15, pp. 839-851, 2020, doi: 10.1109/TIFS.2019.2922229.
[5] Dice, L.R. 1945. “Measures of the amount of ecologic associationbetween species.” Ecology, Vol. 26: pp. 297–302.
Figures