4064
Variational Model Augmented Deep Learning for Small Training Data MRI Thigh Muscle Segmentation1Mathematics, Applied Mathematics and Statistics, Case Western Reserve University, Cleveland, OH, United States, 2Lerner Research Institute, Cleveland Clinic, Cleveland, OH, United States
Synopsis
Keywords: Muscle, Machine Learning/Artificial Intelligence, muscle segmentation, fusion, variational model
Accurate automatic MRI thigh muscle group segmentation is essential for muscle morphology and composition which are related to osteoarthritis and sarcopenia. Due to challenges caused by lack of contrast between muscle groups, Deep Learning (DL) becomes a more natural choice than traditional model based approaches. It is however expensive to obtain a large amount of training data and DL using small training data often results in overfitting. We use a variational model based segmentation method in conjunction with a Bayesian neural network to optimize the train framework, producing about 1.6% increase in dice coefficients while working with minimal annotated data.Introduction
Thigh muscle morphology and composition obtained from MRI have been suggested as potential imaging biomarkers for multiple diseases including osteoarthritis and sarcopenia. Manual segmentation is time consuming and is prone to intra- and inter-operator variations. In order to receive efficient and reliable quantification of thigh muscles, robust and fast reproducible segmentation methods are desirable. Automatic segmentation of thigh muscles faces several challenges such as the quality of MRI scans and the physiology of thigh muscles itself, including tightly bundled sub muscle groups with no clear boundaries between them. This makes automatic semantic segmentation based on just image intensity literally impossible. Pure DL methods need a large amount of training data1. In work 2, a semi-automatic approach requiring users to draw markers and anti-markers for every input data set was proposed. Our proposed framework seeks to combine the benefits of deep learning and variational models for an automatic scheme. Our goal is to reach optimal results with minimal annotated training data.Methods
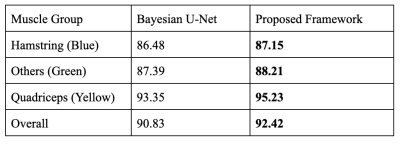
High-resolution T1-weighted turbo spin-echo (TSE) MRI data collected from 23 subjects (age 35.4±6.8, BMI 25.3±4.1, 12 Male) using a 3T MRI scanner with an 18-channel flex coil and anterior/posterior receive arrays for bilateral thigh imaging at 10 years after anterior cruciate ligament reconstruction were used for this study. The sequence parameters include TR/TE = 607-795/10 (ms), an FOV of 400x312x140 mm3 , and with matrix size (432-512)x(383-432)x28. The calculation was done on each slice after each image was interpolated to an image size of 512x512. The three muscle groups annotated are the quadriceps, hamstrings and other muscles (adductor group, gracilis and sartorius).We demonstrate the proposed hybrid scheme using a specific variational model3 and Bayesian U-Net4,5. The framework itself is however generic and can be used to enhance any neural network performance. Starting from a small training data with ground truth (GT) from 5 subjects as shown in Figure 1, Bayesian U-Net is able to obtain an average Dice coefficient of 90.83%. We aim to further increase the accuracy. The strategy is to use the variational method1 to obtain initial results of some of the other subjects (say 10) and treat them as non-perfect “ground truth” GT2. Model3 makes use of a simultaneous registration and segmentation algorithm that has proved to be effective in the segmentation of thigh muscles. It results in minimizing an energy function consisting of 3 terms -- segmentation, cross entropy and registration: E(Θ,u,T) = Eseg(Θ,u) +α ECE (u,s°T) +β Ereg(Θ,T) with u, T, Θ the segmentation map, deformation field and model parameters and α, β balancing parameters. The segmentation term is intensity based and focuses on segmenting the muscles as a whole from the others. The registration term registers a template with ground truth segmentation to the new input and the registered segmentation serves as a shape prior that is adopted to the model using cross entropy. These results have Dice coefficients about 85~90%. We alternate Bayesian U-Net training using the true ground truth GT and the non-perfect ground truth GT2 respectively in an iterative way until convergence. Note instead of combining GT and GT2 in one network, we use them in two networks with different frequencies in a sequential way so that the parameters learned from the previous network serves as the initial of the next network. In our experiments, we train 4 times more often using GT than GT2 in terms of epochs. The process is repeated a few times. This allows the network to pick up new patterns from GT2 while getting penalized for wrong patterns it learns from GT2.
We use Bayesian U-Net as the backbone network. It is a modification of the U-Net that can quantify the uncertainty of the predicted segmentation map at each pixel. It calculates this using Monte Carlo parameter dropouts to generate different predictions from the same trained network at evaluation. Mean values and variances are then calculated and serve as the prediction and uncertainty quantification.
Results
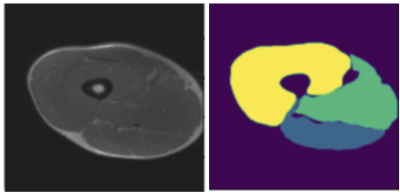
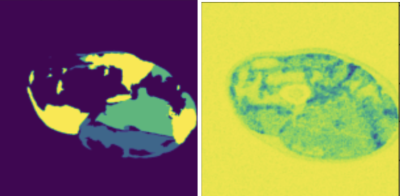
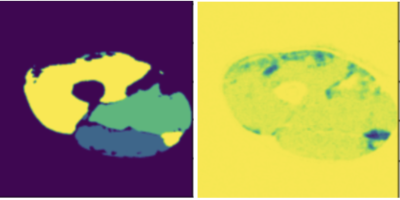
We compare the proposed framework with Bayesian U-Net. Figure 2 shows one example of the thigh muscle MRI slice and the corresponding manual annotation of the 3 muscle groups.The improvement in accuracy is highlighted in Figure 1 for quantitative interpretations and in Figures 2-4 for qualitative comparison. The segmentation performance is improved across all 3 muscle groups. In particular, most muscle regions being misclassified as background with high certainty using the Bayesian U-Net only approach in Figure 3 are now correctly classified as shown in Figure 4. One can notice that the uncertainty is lower on average and the misclassified regions are being accurately picked up by the uncertainty map in Figure 4 in comparison to Figure 3.Discussion
We plan to select subjects from our dataset that have the closest Weierstrass distance from the annotated ground truth serving as template for registration. This will help improve the accuracy of GT2. Furthermore, we will tune for an appropriate ratio of training on GT and GT2 using the uncertainty map coming from the Bayesian network.Conclusion
By fusing the power of model based algorithms with deep neural networks, we were able to improve thigh muscle segmentation on MR images using small training dataset.Acknowledgements
NIH/NIAMS R01 AR075422.References
1. Kemnitz J, Baumgartner CF, Eckstein F et al. Clinical evaluation of fully automated thigh muscle and adipose tissue segmentation using a U-Net deep learning architecture in context of osteoarthritic kneepain. Magnetic Resonance Materials in Physics, 33(4):483-493, 2020.
2. Guo W, Judkovich M, Lartey R, Xie D, Yang M, Li X, “A Marker Controlled Active Contour Model for Thigh Muscle Segmentation in MR Images”, International Society for Magnetic Resonance in Medicine Conference, 2021.
3. Li H , Guo W, Liu J, Cui L and Xie D, Image Segmentation with Adaptive Spatial Priors from Joint Registration, SIAM Journal on Imaging Sciences, Vol.15,No.3, pp.1314-1344, 2022.
4. Gal Y and Ghahramani Z. “Dropout as a Bayesian Approximation: RepresentingModel Uncertainty in Deep Learning”. In: (2015). doi: 10.48550/ARXIV.1506.02142. url:https://arxiv.org/abs/1506.02142.
5. Hiasa Y, Otake Y, Takao M, Ogawa T, Sugano N, and Sato Y. “Automated Muscle Segmentation from Clinical CT using Bayesian U-Net for Personalized Musculoskeletal Modeling”. In: (2019).
Figures