4061
Feasibility of Automated Segmentation of 3D Shoulder Muscle Volume via Deep Learning for Rotator Cuff Repair Patients
Mingrui Yang1, Bong-Jae Jun1, Tammy Owings1, Joshua Polster1, Carl Winalski1, Kathleen Derwin1, Eric Ricchetti1, and Xiaojuan Li1
1Cleveland Clinic, Cleveland, OH, United States
1Cleveland Clinic, Cleveland, OH, United States
Synopsis
Keywords: Muscle, Machine Learning/Artificial Intelligence
It has been shown that muscle volume and fat fraction play significant roles in musculoskeletal disorder diagnosis and prognosis. Reliable clinical tools for their evaluation, however, are currently missing. One hurdle is the challenging and laborious manual segmentation process on MR images. We proposed here a deep learning based automated tool for 3D shoulder muscle volume segmentation and achieved accurate segmentation results on clinical MR images from rotator cuff repair patients. The proposed model can be a valuable tool for shoulder muscle volume quantification and subsequent fat fraction analysis to further understand their association with clinical outcomes following shoulder procedures.Introduction
Increasing evidence has been shown from recent clinical and basic science studies that muscle volume and fat fraction play significant roles in diagnosis and treatment prognosis of shoulder disorders. For instance, muscle atrophy and fatty infiltration have been shown to be significantly correlated with worse clinical outcomes after rotator cuff repair (RCR) 1,2. Reliable tools to quantify muscle volume and fat fraction will provide physicians powerful methods to optimize patient management and improve patient outcomes, since muscle pathology is potentially modifiable (with exercise training or pharmacological interventions) and can impact surgical decision-making, including the type of surgical repair chosen. Such tools, however, are currently missing because these quantitative measures heavily depend on accurate muscle segmentation, which is extremely challenging and laborious, and subject to intra- and inter-reader variation. This study aims to build an automated shoulder muscle segmentation model from clinical MR images using deep learning as the first step towards building reliable tools for muscle volume and fat fraction quantification.Methods
Preoperative clinical MR images of 24 rotator cuff repair patients (10 partial tear, 14 full thickness tear) were retrieved from the Cleveland Clinic Orthopaedic Outcomes (OME) cohort, a prospective orthopaedic surgical cohort within the Cleveland Clinic health care system. In addition, clinical MR images from 10 patients with no tear were retrieved. The MR images were collected with various T1-weighted sequences on different MR scanners (Siemens Healthcare, Erlangen, Germany) from different locations, with matrix size varying from 256 to 512, in-plane resolution varying from 0.23mm to 0.86mm, and slice thickness varying from 3mm to 6mm. The collected MR images were graded by musculoskeletal radiologists in routine clinical practice. All experiments were approved by the institutional review board. Three shoulder muscle groups (supraspinatus, combined infraspinatus / teres minor, and subscapularis) were manually segmented for 3D muscle volumes by a trained reader. The 34 manually segmented cases were randomly split into 21/7/6, resulting in 304/103/74 slices for deep learning model training, validation, and testing, respectively. The MR images were preprocessed with contrast limited adaptive histogram equalization (CLAHE) for better image contrast and homogeneity, and saved into 16-bit unsigned integer format. The images were further augmented with random rotation, translation, and zooming before fed into the deep learning model for training. The deep learning model architecture was based on the DeepLabV3+ 3 with ResNet50 as the backbone. Categorical cross entropy was used as the loss function. The model was pretrained on clinical shoulder CT images and further trained on MR images for 1000 epochs using the ADAM optimizer with a learning rate of 1e-5 and batch size of 16. The training, validation, and test of the models were realized with the Python deep learning framework Tensorflow with the Keras API on a NVIDIA V100s (32GB) GPU. The Dice coefficients (DC), the average symmetric surface distance (ASSD), the 95 percentile Hausdorff distance (HD95), and the relative absolute volume difference (RAVD), all calculated in 3D, were used as metrics for model evaluation. The model performance was compared on the test set against the well-known 2D U-Net 4.Results
The benefits of applying CLAHE were shown in Figure 1, where the CLAHE results clearly had better brightness and more homogeneous contrast across the images, which helped with the model training. The proposed model achieved a mean Dice of 0.933, a mean ASSD of 0.289 mm, a mean HD95 of 1.104 mm, and a mean RAVD of 3.6% on the test set. In comparison, the benchmark 2D U-Net had a mean Dice of 0.629, a mean ASSD of 1.641 mm, a mean HD95 of 9.717 mm, and a mean RAVD of 42.1%. Figure 2 depicted the performance of the proposed model in overall and compartment-wise 3D Dice coefficients in violin plots for the 6 test cases. Segmentation examples of the 3 muscle groups of a test case at 3 slice positions (most medial, central, and most lateral) were shown in Figure 3.Discussion
Few studies have investigated automated segmentation model for 3D shoulder muscle volume on clinical MR images for RCR patients. In this study we developed a deep learning model to achieve this task. The proposed model outperformed the 2D U-Net model in all 4 evaluation metrics utilized in the study. It’s worth noting that the model performed worse at the edge slices than at the central slices, which may be due to one of the limitations of this study -- the relatively small number of manually labelled slices available for model training, validation and testing, though data augmentation was applied. Future work will increase the number of manually segmented cases to further improve the model performance.Conclusion
This study demonstrated the feasibility of an accurate and reliable automated shoulder muscle segmentation model for clinical MR images based on deep learning that will substantially reduce the time required for 3D muscle volume and quality analysis. The proposed model can provide a valuable tool for quantifying 3D shoulder muscle volume and subsequent fat fraction analysis to further understand their association with clinical outcomes following shoulder procedures such as rotator cuff repair.Acknowledgements
NIH/NIAMS R01 AR075286; Cleveland Clinic internal funding: Artificial Intelligence in Medicine.References
- Gladstone JN, Bishop JY, Lo IK, Flatow EL. Fatty infiltration and atrophy of the rotator cuff do not improve after rotator cuff repair and correlate with poor functional outcome. AmJSports Med. 2007;35(5):719-728.
- Gerber C, Schneeberger AG, Hoppeler H, Meyer DC. Correlation of atrophy and fatty infiltration on strength and integrity of rotator cuff repairs: a study in thirteen patients. J Shoulder Elbow Surg. 2007;16(6):691-696.
- Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y (eds). Computer Vision – ECCV 2018. ECCV 2018. Lecture Notes in Computer Science(), vol 11211. Springer, Cham. https://doi.org/10.1007/978-3-030-01234-2_49
- Ronneberger OF, Philipp; Brox, Thomas. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv. 2015:1505.04597
Figures
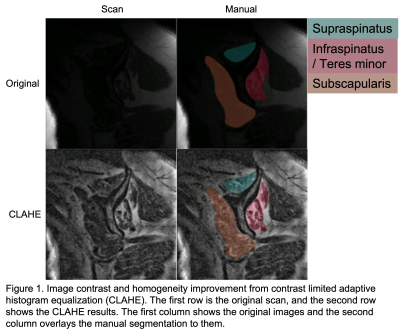
Image
contrast and homogeneity improvement from contrast limited adaptive histogram
equalization (CLAHE). The first row is the original scan, and the second row shows the CLAHE results. The first column
shows the original images and
the second column overlays the manual segmentation to them.
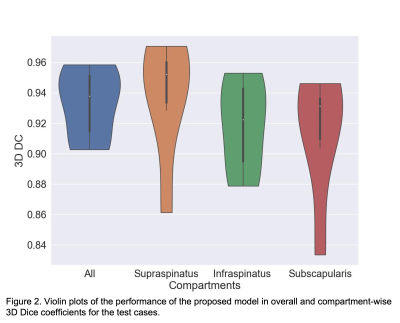
Violin
plots of the performance of the proposed model in overall and compartment-wise
3D Dice coefficients for the test cases.
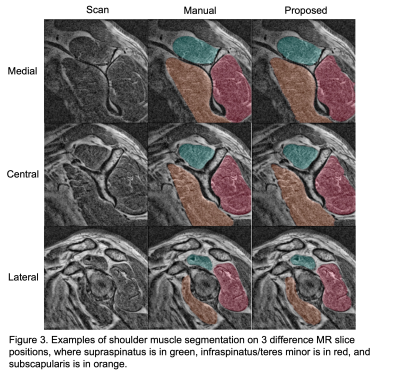
Examples
of shoulder muscle segmentation on 3 difference MR slice positions, where
supraspinatus is in green, infraspinatus/teres minor is in red, and
subscapularis is in orange.
DOI: https://doi.org/10.58530/2023/4061