4035
Exploring reproducibility in deep learning-based parallel imaging reconstruction1Seoul National University, Seoul, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
The performance of a deep neural network can be affected by software and hardware setups when training the network and, therefore, can vary from training to training. This issue of reproducibility, which can be referred to as an “intrinsic” reproducibility of deep learning, can be critical for academic research because reproducibility is a key requirement for journal papers. In this study, we explore this intrinsic reproducibility issue for deep learning-powered parallel imaging reconstruction by using a popular end-to-end variational network. This study may provide minimal requirements for reproducible research in network training.Introduction
In recent years, deep learning has shown improved performance in various MRI reconstruction tasks including parallel imaging1, quantitative susceptibility mapping2, and image denoising3. Despite its success, deep learning-based reconstruction still suffers from unexpected issues which, in many cases, are not problems in conventional algorithms. One of the important issues, which was introduced recently, is “intrinsic” reproducibility issue in deep learning4, where network performances are different for each training trial due to details in software and hardware setups (Figure 1). This issue implies that current network training may not guarantee reproducibility which is an important issue for academic research. Particularly, this issue can be critical when comparing the performances of various networks, because the evaluation results may differ from those of the original networks if the networks are re-trained.In this study, we explored the intrinsic reproducibility issue in deep learning-based parallel imaging reconstruction using a popular end-to-end variational network5 (abbreviated in E2E-VarNet) as a representative deep learning model. By investigating sources affecting reproducibility and their effects on network performance, this study may provide guidelines for performing reproducible network training.
Methods
For network training and evaluation, T1-weighted brain images from FastMRI challenge dataset6, which contains a total of 5268 slices of full-sampled k-space data from 340 subjects, were used. From the full-sampled k-space data, under-sampled data were retrospectively generated using a k-space uniform sampling mask with acceleration factor = 4. Finally, a total of 5268 pairs of under-sampled data and full-sampled data were generated and used for input and label pairs for deep learning. The dataset was divided into 3400, 1238, and 630 pairs for generating training, validation, and test dataset, respectively. Furthermore, the test dataset was refined to exclude the data with large image artifacts, resulting in a total of 566 pairs for the test dataset. Using the training and validation datasets, E2E-VarNet networks were trained. The E2E-VarNet networks were implemented based on public code available at the following link: https://github.com/facebookresearch/fastMRI. The training loss was set to 1 – SSIM (Structural Similarity Index) to maximize SSIM. The training was stopped in epoch 45, and the final network was chosen to have the best validation loss.For reproducible network training, an additional experimental setup was performed referring to the previous work4. The network was trained in the same computational infrastructure equipped with i9-7800X CPU (Intel, Santa Clara, CA, USA) and Titan XP GPU (NVIDIA, Santa Clara, CA, USA). The training code was implemented using PyTorch7 and contains a CUDA setup which set determinism option on and benchmarking option off8. Also, mini-batch order and initial network weights were set to the constant. This experimental setup was set to baseline setup and the training reproducibility was verified by comparing training losses in all training steps for two different training trials.
After confirming the training reproducibility, we trained networks by changing each following source: CUDA option either determinism or benchmarking, mini-batch order, and initial network weight. Then, network performances were compared to verify that each source affects the network performance. The network performances were evaluated by SSIM, PSNR (Peak Signal to Noise Ratio), and NMSE (Normalized Mean Squared Error) values in the test dataset. For a statistical test, a paired t-test with Bonferroni correction was performed.
Results
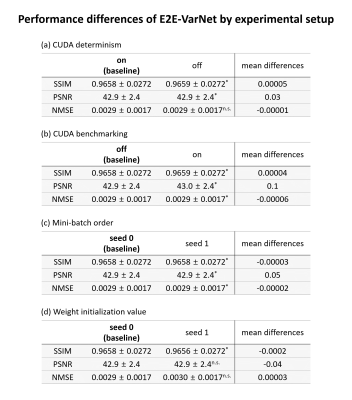
When the network was trained with a baseline experimental setup, it was confirmed that network training was reproducible, reporting the same loss values in all training steps. When either CUDA determinism or benchmarking option differed from the baseline, network performance yielded different performances, violating reproducibility (Fig. 2a-b). Furthermore, the mini-batch order and the initial network weight also affected the training reproducibility, resulting in performance differences (Fig. 2c-d). The statistical test via the paired t-test confirmed that performance differences were statistically significant (p-value < 0.05/3). However, the differences were very small (0.0002 for SSIM, 0.1 for PSNR, and less than 0.0001 for NMSE).Discussion and Conclusion
In this work, we explored the intrinsic reproducibility issue in E2E-VarNet. Four sources were tested to affect the training reproducibility, implying that these sources need to be considered for reproducible network training. However, the performance variance of E2E-VarNet was small compared to that of the other network such as QSMnet2 (SSIM differences were up to 0.005; reported in a previous work9), which is a deep neural network for quantitative susceptibility mapping. The difference in performance variance between E2E-VarNet and QSMnet may be related to differences in training dataset size (300 subjects for E2E-VarNet and 7 subjects for QSMnet), network structure (i.e. unrolled network for E2E-VarNet and U-net for QSMnet), and tasks (i.e. parallel imaging vs. quantitative susceptibility mapping).When reporting the detailed experimental setup including these sources, we recommend sharing the training code because CUDA options, mini-batch order, and initial network weight can be controlled by adding several lines in codes and setting constant random seeds.
Acknowledgements
This work has been supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF-2021R1A2B5B03002783, NRF-2019M3C7A1031994)References
1. Hammernik, K. et al. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 79, 3055–3071. (2018).
2. Yoon, J. et al. Quantitative susceptibility mapping using deep neural network: QSMnet. Neuroimage 179, 199–206. (2018).
3. Manjón, José V., and Pierrick Coupé. MRI denoising using deep learning. International Workshop on Patch-based Techniques in Medical Imaging, Springer, Cham. (2018).
4. Chungseok. Oh. et al. Intrinsic reproducibility issues in deep learning-based MR reconstruction. Proceedings of the International Society for Magnetic Resonance in Medicine, 6117. (2022).
5. Sriram, Anuroop, et al. End-to-end variational networks for accelerated MRI reconstruction. International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, Cham, (2020).
6. Zbontar, Jure, et al. fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:1811.08839. (2018).
7. Paszke, Adam, et al. Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems 32. (2019).
8. Reproducibility — PyTorch 1.13 documentation. https://pytorch.org/docs/stable/notes/randomness.html. (2022).
9. Chungseok. Oh. et al. Fair comparison in deep learning QSM. Joint Workshop on MR phase, magnetic susceptibility and electrical properties mapping. Lucca, Italy. (2022).
Figures