4004
Ungated and free-breathing radial simultaneous multi-slice cardiac T1 mapping1Biomedical Engineering, University of Utah, Salt Lake City, UT, United States, 2Utah Center for Advanced Imaging Research, University of Utah, Salt Lake City, UT, United States, 3Cardiology, University of Utah, Salt Lake City, UT, United States
Synopsis
Keywords: Myocardium, Relaxometry
Cardiac T1 mapping has been shown to be a promising method for assessing different cardiomyopathies. Most cardiac T1 mapping methods require ECG gating with long breath holds to capture specific desired cardiac phases and to minimize breathing motion. However, certain cardiomyopathies can make it difficult for patients to maintain a breath hold for the duration of a cardiac T1 mapping sequence. Furthermore, some of these diseases, for example atrial fibrillation with changing R-R intervals can make capturing a specific cardiac phase difficult. Here we propose a radial simultaneous multi-slice (SMS) cardiac T1 mapping sequence without ECG gating or breath holding.Introduction
Cardiac T1 mapping has shown promise in differentiating various cardiomyopathies [1-5]. The 11 heartbeat MOLLI sequences [6] are among the most popular T1 mapping sequences due to their high precision. The 9 heartbeat shMOLLI [7] and 10 heartbeat SASHA [8] are shorter sequences that reduce the breath-hold burden; however, these sequences still rely on ECG gating for accurate T1 mapping. Other works using radial single-slice inversion recovery-based sequences have been proposed to accelerate T1 acquisitions [9-11], but still rely on accurate ECG gating. For patients with atrial fibrillation, the irregularity in the ECG signal can result in unusable cardiac T1 maps. As such, there is an unmet need for a T1 mapping framework without ECG gating or breath-holding. MR Fingerprinting and multi-tasking methods that do not rely on ECG-gating and simultaneously estimate multiple parameters have been proposed [12, 13]. Here, we develop a radial simultaneous multi-slice (SMS) dedicated T1 mapping sequence without ECG gating or breath-holding using a subspace constrained reconstruction and model-based regularization for accurate T1 mapping.Methods
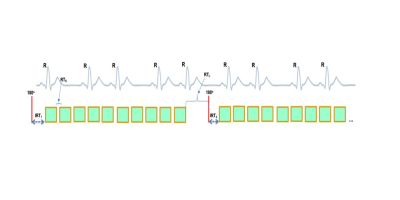
Data AcquisitionIn the proposed ungated T1 mapping framework, an inversion pulse is followed by the acquisition of five images with a short recovery time spacing of $$$RT_s$$$ between each image and an inversion recovery time of $$$IRT_1$$$. After a long recovery time of $$$RT_L$$$, another five images with $$$RT_s$$$ spacing between each image and an inversion recovery time of $$$IRT_2$$$ are acquired for an increased sampling of the T1 recovery curve. Figure 1 demonstrates a schematic of the ungated T1 mapping sequence. The acquisition parameters were $$$TR=2.50ms$$$, $$$TE=1.29ms$$$, $$$FA=8^{\circ}$$$, $$$FOV=320mm^2$$$, $$$RT_s=200ms$$$, $$$RT_L=2500ms$$$, $$$IRT_1=11ms$$$, and $$$IRT_2=100ms$$$.
Self-Gating
Radial SMS data were interpolated onto a cartesian grid using a SMS GRAPPA operator gridding [14] (SMS-GROG) with a sliding window approach in order to generate preliminary images with a temporal resolution of $$$42ms$$$ and a temporal footprint of $$$359ms$$$ for self-gating. Cardiac masks were generated using temporal standard deviation maps for each inversion pulse image group where high-intensity regions will mostly reflect changes in contrast of the blood pool at various inversion times. These temporal standard deviation maps were averaged, smoothed, and then used to generate heart images [15]. To extract the cardiac signal, the heart images were vectorized spatially and principal component analysis was used through time. The highest amplitude basis function among the first ten basis functions within the cardiac motion range of $$$0.5-2.2 Hz$$$ was chosen as the cardiac signal [16].
Image Reconstruction
The subspace constrained reconstruction with model-based regularization was used to jointly reconstruct the T1-weighted images acquired at different inversion times and to estimate the T1 maps [17, 18]. The reconstruction framework is described by the equation below:
$$m=argmin_m \frac{1}{2} \lVert Am-d \rVert ^2_2 + λ_s \lVert \sqrt{ \nabla_x + \nabla_y + ε} \rVert ^2_2 + λ_m \lVert M(m) - \hat{m} \rVert ^2_2$$
$$ s.t. m = U_kV_k$$
where $$$d$$$ is multi-coil cartesian k-space data after interpolation using SMS-GROG. $$$A=φDFS$$$ is the forward encoding matrix which describes the physics of MRI reconstruction. $$$φ$$$ is the phase modulation that combines and separates SMS slices. $$$D$$$ is the under-sampling mask describing the sampling trajectory. $$$F$$$ is the Fourier transform and $$$S$$$ are the coil sensitivities. $$$m$$$ represents the multi-slice images to be reconstructed. $$$U_k$$$ are the first $$$k$$$ temporal basis functions and $$$V_k$$$ are their corresponding image coefficients. $$$λ_s$$$ is the spatial regularization parameters for the spatial total variation constraints. $$$ε$$$ is a small positive constant to avoid singularity. $$$λ_m$$$ is the model-based regularization parameter for the $$$L_2$$$ norm between the estimated image $$$m$$$ that is motion-compensated using $$$M$$$ and the model-based image $$$\hat{m}$$$. $$$\hat{m}$$$ is updated every iteration by generating T1 maps from $$$m$$$ using a T1 fitting pattern recognition algorithm. The motion compensating operator, $$$M$$$, was folded into the iterative reconstruction using the pTV method [19]. The phase of $$$m$$$ and $$$\hat{m}$$$ are kept the same. T1 fitting was performed using a pattern recognition algorithm [11] with a slice profile and phase corrected dictionary consisting of B1 ranging from $$$0.6-1.4$$$ to account for flip angle variations, and T1 ranging from $$$100-2700$$$. This dictionary was also used to generate temporal basis functions for the subspace constrained reconstruction.
Results
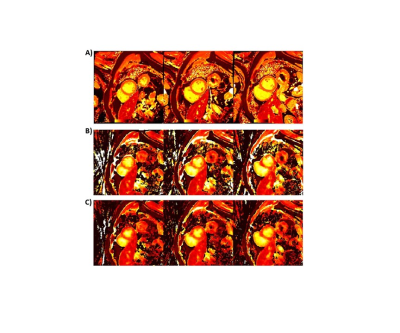
Figure 2 shows the near-diastolic and near-systolic T1 maps obtained from a patient with Transthyretin Amyloidosis with Val30Met mutation on Siemens 3T Prisma scanner using the proposed ungated sequence. Corresponding native T1 maps obtained using the standardCartesian MOLLI-5(3)3 sequence are also shown. The mean and standard deviation of T1 values computed using the 16-segment AHA model [20] for the standard MOLLI T1 maps were $$$1234 \pm 35ms$$$. Corresponding mean diastolic and mean systolic T1 values from the proposed ungated sequence were $$$1203 \pm 44ms$$$ and $$$1145 \pm 90ms$$$ respectively.Conclusions
Here we show the feasibility of a free-breathing ungated T1 mapping approach that uses a fixed set of acquisition parameters with an image-based self-gating approach and a subspace constrained reconstruction with model-based regularization that includes motion compensation to obtain near-systolic and near-diastolic T1 maps from a single acquisition. Additional studies are required to evaluate the performance of the proposed approach in healthy volunteers as well as in patients with cardiomyopathies.Acknowledgements
No acknowledgement found.References
[1] Dall'Armellina E, Piechnik SK, Ferreira VM, Si QL, Robson MD, Francis JM, et al. Cardiovascular magnetic resonance by non contrast T1-mapping allows assessment of severity of injury in acute myocardial infarction. J Cardiovasc Magn Reson 2012;14:15.
[2] Karamitsos TD, Piechnik SK, Banypersad SM, Fontana M, Ntusi NB, Ferreira VM, et al. Noncontrast T1 mapping for the diagnosis of cardiac amyloidosis. JACC Cardiovasc Imaging 2013;6(4):488-97.
[3] Sado DM, White SK, Piechnik SK, Banypersad SM, Treibel T, Captur G, et al. Identification and assessment of Anderson-Fabry disease by cardiovascular magnetic resonance noncontrast myocardial T1 mapping. Circ Cardiovasc Imaging 2013;6(3):392-8.
[4] Taylor AJ, Salerno M, Dharmakumar R, Jerosch-Herold M. T1 Mapping: Basic Techniques and Clinical Applications. JACC Cardiovasc Imaging 2016;9(1):67-81.
[5] Schelbert EB, Messroghli DR. State of the Art: Clinical Applications of Cardiac T1 Mapping. Radiology 2016;278(3):658-76.
[6] Kellman P, Wilson JR, Xue H, Ugander M, Arai AE. Extracellular volume fraction mapping in the myocardium, part 1: evaluation of an automated method. J Cardiovasc Magn Reson 2012;14:63.
[7] Piechnik SK, Ferreira VM, Dall'Armellina E, Cochlin LE, Greiser A, Neubauer S, et al. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson 2010;12:69.
[8] Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn Reson Med 2014;71(6):2082-95.
[9] Wang X, Voit D, Roeloffs V, Uecker M, Frahm J. Fast Interleaved Multislice T1 Mapping: Model-Based Reconstruction of Single-Shot Inversion-Recovery Radial FLASH. Comput Math Methods Med 2018;2018:2560964.
[10] Gensler D, Morchel P, Fidler F, Ritter O, Quick HH, Ladd ME, et al. Myocardial T1: quantification by using an ECG-triggered radial single-shot inversion-recovery MR imaging sequence. Radiology 2015;274(3):879-87.
[11] Marty B, Coppa B, Carlier PG. Fast, precise, and accurate myocardial T1 mapping using a radial MOLLI sequence with FLASH readout. Magn Reson Med 2018;79(3):1387-98.
[12] Lima da Cruz GJ, Velasco C, Lavin B, Jaubert O, Botnar RM, Prieto C. Myocardial T1, T2, T2*, and fat fraction quantification via low-rank motion-corrected cardiac MR fingerprinting. Magn Reson Med 2022;87(6):2757-74.
[13] Cao T, Wang N, Kwan AC, Lee HL, Mao X, Xie Y, et al. Free-breathing, non-ECG, simultaneous myocardial T1 , T2 , T2 *, and fat-fraction mapping with motion-resolved cardiovascular MR multitasking. Magn Reson Med 2022;88(4):1748-63.
[14] Tian Y, Mendes J, Pedgaonkar A, Ibrahim M, Jensen L, Schroeder JD, et al. Feasibility of multiple-view myocardial perfusion MRI using radial simultaneous multi-slice acquisitions. PLoS One 2019;14(2):e0211738.
[15] Tian Y, Mendes J, Wilson B, Ross A, Ranjan R, DiBella E, et al. Whole-heart, ungated, free-breathing, cardiac-phase-resolved myocardial perfusion MRI by using Continuous Radial Interleaved simultaneous Multi-slice acquisitions at sPoiled steady-state (CRIMP). Magn Reson Med 2020;84(6):3071-87.
[16] Zhou R, Weller DS, Yang Y, Wang J, Jeelani H, Mugler JP, 3rd, et al. Dual-excitation flip-angle simultaneous cine and T1 mapping using spiral acquisition with respiratory and cardiac self-gating. Magn Reson Med 2021;86(1):82-96.
[17] Feng L, Wen Q, Huang C, Tong A, Liu F, Chandarana H. GRASP-Pro: imProving GRASP DCE-MRI through self-calibrating subspace-modeling and contrast phase automation. Magn Reson Med 2020;83(1):94-108.
[18] Adluru G, McGann C, Speier P, Kholmovski EG, Shaaban A, Dibella EV. Acquisition and reconstruction of undersampled radial data for myocardial perfusion magnetic resonance imaging. J Magn Reson Imaging 2009;29(2):466-73.
[19] Vishnevskiy V, Gass T, Szekely G, Tanner C, Goksel O. Isotropic Total Variation Regularization of Displacements in Parametric Image Registration. IEEE Trans Med Imaging 2017;36(2):385-95.
[20] Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Int J Cardiovasc Imaging 2002;18(1):539-42.
Figures