3989
Application of natural language processing to post-structuring of rectal cancer MRI reports1Beijing Aviation General Hospital, Beijing, China
Synopsis
Keywords: Pelvis, Cancer
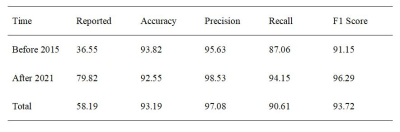
We applied natural language processing (NLP) to the extraction of relevant information from MRI reports of rectal cancer written in Chinese. We used 358 MRI reports written between 2015 and 2021 to develop a rule-based NLP model to extract 11 key image features. The accuracy, precision, recall, and F1 score of our NLP model for correct extraction of values from reports were 93.82%, 95.63%, 87.06%, and 91.15% for pre-2015 reports, and 92.55%, 98.53%, 94.15%, and 96.29% for post-2021 reports. Our NLP system with rule-based pattern matching realized the rapid and accurate structured processing of rectal cancer MRI reports.Introduction
A magnetic resonance imaging (MRI) pelvic scan is necessary for the local staging of rectal cancer because of the need for high-resolution imaging of anatomic details to determine local tumor extension 1. Although existing guidelines and medical consensus2-4 recommend the use of structured reports, such reports are not widely used because they cannot represent all the semantic information expressed in natural language. Therefore, a large number of image reports in the medical record system are still stored in the form of free-form text. This free-form text format is the main obstacle to the rapid extraction of information for use by clinicians, researchers, and health care information systems. As a result, most image reports are only checked once by clinicians, are saved in the medical records system in the form of the original free-form text, and are rarely used for data mining. Natural language processing (NLP) is an important aspect of computer science and artificial intelligence. It involves the study of various theories and methods for realizing effective communication between humans and computers using natural language. The post structuring of image reports refers to the extraction of structured information from text-based unstructured data written by radiologists. Assuming that all relevant imaging features were recorded in the report, NLP should be able to extract the structured information from the report that is required to determine the index lesions5.Methods
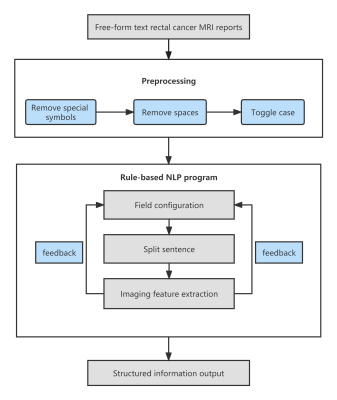
A rule-based NLP model that could extract 11 key image features of rectal cancer was constructed using 358 reports of rectal cancer MRI written between 2015 and 2021. Example MRI reports are shown in Figure 1. Fifty reports written before 2015 and 50 written after 2021 were used as test datasets, and the reference standard was determined by manual extraction of information by two radiologists. Table 1 lists the imaging descriptors to be extracted from the structured fields. The four structured procedures in the NLP program are outlined in Figure 2, and were: field configuration, splitting of sentences, field value extraction, and feedback modification. The length and reporting rate of image features in pre-2015 and post-2021 datasets, as well as the accuracy, precision, recall, and F1 score of feature extraction by the NLP system, were compared. The time required for the NLP to extract data was compared with that required by the radiologists.Results
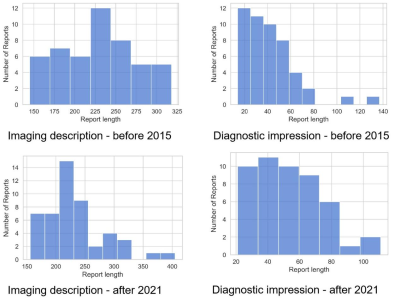
Figure 3 shows the word count in the descriptions of colorectal cancer in MRI reports before 2015 and after 2021. There was no significant difference in the number of imaging description words between the two datasets (p=0.879), although there was a significant difference in the number of diagnostic impression words (p=0.021). The performance of the NLP system in the post-structuring of rectal cancer MRI reports is shown in Table 2. The average time for the manual reviewers to extract the imaging features was 1.31 ± 0.68 minutes per report, while the NLP generated the total results in less than 1 second.Discussion
Our approach utilized a rule-based method that identified text referring to tumor and its corresponding features in rectal MRI reports. Liu et al. developed a rule-based NLP system that successfully extracted nine imaging features and their values from all lesions, and which showed recall and precision of 91.0% and 92.6%, respectively[11]. Rule-based pattern matching uses regular expressions to represent the string patterns that need to be matched. It has good flexibility for natural language texts and the extraction process is simple and easy to operate, although the extraction effectiveness is highly dependent on the formulated rules (string patterns), making it most suitable for expressing standardized texts. In rule-based pattern matching, the term “regular expression” refers to the algebraic expression used to describe the regular set. It is a text pattern composed of particular characters such as ordinary characters and wildcards, and is commonly used in the search for text content. The regular expression can match the text according to an algorithm to realize the function of extracting sub-character strings from a string, and is the basis of the rule-based pattern-matching text information extraction method[14]. Because the MRI reports of rectal cancer at our center were single-disease reports and had a certain standardization, the text structure and data characteristics could be quickly sorted out. Our rule-based pattern-matching method is suitable for structured processing to realize structured information extraction of unstructured text. Since almost all MRI reports of rectal cancer after 2021 used structured templates, the performance of the rule-based NLP system for image feature extraction was higher for reports after 2021 than for reports before 2015. Such research could promote the writing standard of single center imaging reports.Conclusions
Our NLP system with rule-based pattern matching realized the rapid and accurate structured processing of rectal cancer MRI reports. MRI reports with structured templates are more suitable for NLP-based extraction of information.Acknowledgements
This work has been supported by National Natural Science Foundation: 82202258.References
1 Fowler KJ, Kaur H, Cash BD et al (2017) ACR Appropriateness Criteria ® Pretreatment Staging of Colorectal Cancer. Journal of the American College of Radiology 14:S234-S244
2 Nougaret S, Rousset P, Gormly K et al (2022) Structured and shared MRI staging lexicon and report of rectal cancer: A consensus proposal by the French Radiology Group (GRERCAR) and Surgical Group (GRECCAR) for rectal cancer. Diagnostic and Interventional Imaging 103:127-141
3 Alvfeldt G, Aspelin P, Blomqvist L, Sellberg N (2021) Radiology reporting in rectal cancer using MRI: adherence to national template for structured reporting. Acta Radiologica:1286487060
4 National HCOT (2020) National guidelines for diagnosis and treatment of colorectal cancer 2020 in China (English version). Chin J Cancer Res 32:415-445
5 Liu Y, Liu Q, Han C, Zhang X, Wang X (2019) The implementation of natural language processing to extract index lesions from breast magnetic resonance imaging reports. BMC Med Inform Decis Mak 19:288
6 Liu Y, Liu Q, Han C, Zhang X, Wang X (2019) The implementation of natural language processing to extract index lesions from breast magnetic resonance imaging reports. BMC Med Inform Decis Mak 19:288
7 Thompson K (1968) Programming Techniques: Regular expression search algorithm. Communications of the Acm 11:419-422
Figures