3943
Optimization and comparison of coil combination and spectral registration strategies for in-vivo DW-MRS1College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 2Siemens Healthcare Ltd, Shanghai, China
Synopsis
Keywords: Data Processing, Spectroscopy, Spectra Registration
Diffusion-weighted MRS (DW-MRS) suffers from low signal intensity and large phase errors when applying strong diffusion weighting. Coil combination and spectral registration are vital as spectra with high b-values are usually poorly reconstructed. This work aims at the optimization of post-processing pipeline with singular value decomposition (SVD)-based spectral registration and the comparison of different regimes of coil combination and spectral registration for in-vivo DW-MRS post-processing. The SNR of post-processed spectra and the metabolite diffusivities are assessed. The statistical outcomes suggest that the SVD-based spectral registration could improve the SNR of DW-MRS signal at high b-values and therefore alleviate diffusivity overestimations.Introduction
DW-MRS allows for studying molecular diffusivity to reveal the microstructural and microenvironmental properties in vivo. However, the phase/frequency errors among multiple acquisitions in DW-MRS are non-negligible, especially at large b-values. Many efforts have been made in the past to improve MRS post-processing for a better SNR in the coil combination and spectral registration process. The MRS data processing toolkit FID-A1, GANNET2 and Osprey3 all adopted the time-domain coil combination which calculates the phase and amplitude weighting using the first time point data of FID4,5 (referred to as tdCoilComb). While an SVD-based method was proposed for coil combination6 (svdCoilComb) without referencing to any specific time point. For spectral registration, the robust spectral registration (rSR) method was proposed to align individual transients using an optimal reference and nonlinear least-squares optimization algorithm7. In this work, we proposed an SVD-based spectral registration (svdSR) to improve the quality for DW-MRS. This work compared four different regimes for DW-MRS post-processing. The spectral SNR at different b-values and the apparent diffusion coefficients (ADCs) of different metabolites were assessed.Methods
Instrument:Experiments were performed on a 3T Siemens MAGNETOM Prisma MRI scanner, equipped with 80mT/m gradients and 64-channel head phased-array receive coils.In vivo Experiment: Five healthy volunteers (male/female=1/4; 22-25 years old) participated in this study. The DW-MRS acquisitions were performed using a diffusion-weighted STEAM sequence (TE/TM/TR=62/45.6/1500 ms) in the frontal lobe. The scan parameters included: voxel size = 24*60*25 mm3; spectral width = 4 kHz and 2176 complex datapoints. Diffusion-weighting was applied in three orthogonal directions with diffusion gradient duration (δ) = 20 ms, diffusion time (Δ) = 80 ms. Seven b-values ranging from 0 to 5000 s/mm2 (0, 833, 1667, 2500, 3334, 5000) with 20 shots were acquired.
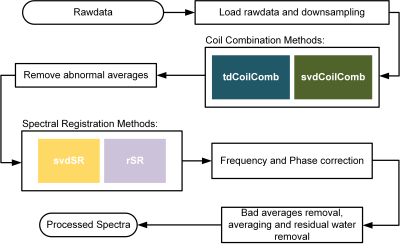
Data Processing:The schematic diagram of DW-MRS processing pipeline was shown in Figure 1. The raw data were reconfigured by down-sampling in the time domain. Two coil combination methods, tdCoilComb4 and svdCoilComb6 were used to combine the individual channels with amplitude and phase weighting respectively.
For spectral registration, we proposed a method based on SVD algorithm, which extracts the principle component to produce the aligned signal in all transients according to the algorithms described below: $${{S}_{unaligned}}=U\Sigma {{V}^{H}}$$$${{S}_{aligned}}={{\Sigma }_{1,1}}{{u}_{n,1}}\left| v_{m,1}^{H} \right|\times {{exp}({\phi {{({{v}_{m,1}})}_{median}}})}$$
where the unaligned signal matrix is denoted as $$${{S}_{unaligned}}\in {{\mathbb{C}}^{n\times m}}$$$; the numbers of complex points and shots are denoted as $$$n, m$$$; $$$H$$$ is the Hermitian transpose. After SVD decomposition, the signal were aligned as product of the maximum singular value $$${{\Sigma }_{1,1}}$$$ and the corresponding left singular vectors $$${{u}_{n,1}}$$$weighted by the amplitude and median phase of right singular vectors $$${{v}_{m,1}}$$$.
At the same time, the rSR method7 was adopted as a comparison in the process of frequency and phase correction before the final averaging step. The averaged spectra was then processed for spectral fitting and ADC calculations. AMARES (jMRUI-v6) algorithm was used to quantify metabolite signals. The metabolite ADCs were calculated from a mono-exponential model (fits up to b4 and b6). The spectral SNR was calculated according to the NAA signal over the standard deviation of spectral noise.
Statistical Analysis:The paired t-test was applied to assess four different regimes for the spectral SNRs and ADC measurements. P < 0.05 was considered statistically significant.
Results
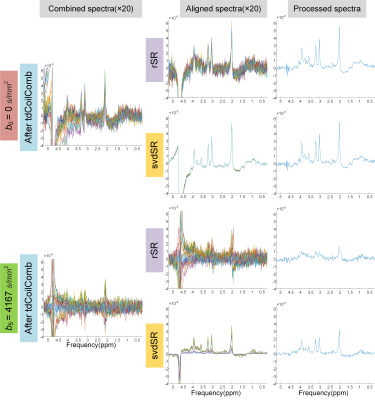
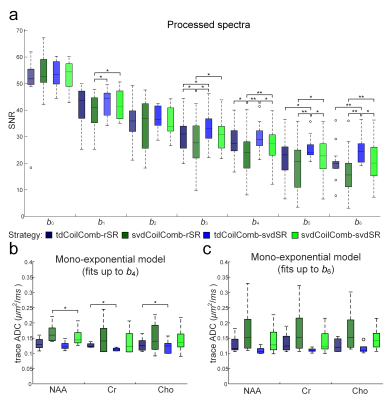
Figure 2 presented the alignment using svdSR and rSR after tdCoilComb at b0 and b5. At b5, the svdSR method showed better performance in recovering the highly diffusion-weighted signals.Figure 3a showed the spectral SNRs of all the post-processed spectra at 7 b-values in 3 directions from five volunteers. The SNR decreased globally as the b-value increased. The SNRs were similar between coil combination methods (tdCoilComb vs. svdCoilComb) while were slightly but significantly differed between spectral registration methods, that was, svdSR method showed a better SNR than rSR at higher b-values, especially when b > 3000 s/mm2. Figure 3b,c presented the trace ADC (arithmetic mean of three directions’ ADCs) of 3 metabolites with different b-ranges in five volunteers. Among four proposed regimes, the trace ADCs calculated from the post-processing using time domain coil combination (tdCoilComb) plus SVD-based spectral registration (svdSR) were the lowest among all pipelines.
Discussion
In this study, SVD-based spectral registration was found to be beneficial to improve in-vivo DW-MRS measurement. The spectra registration using SVD extracts the major component in all spectra, which achieves the denoising and phase/frequency alignment of individual spectra at the same time. However, it remains unclear whether multiple SVD procedures or different numbers of major components extracted would influence the ADC calculation - which needed to be evaluated in the future. Moreover, there are some other coil combine methods such as WSVD8, GLS9, as well as other spectral registration methods such as tdSR10, RATS11, which were also used for MRS post-processing. In the future, these methods could all be parallelly compared and assessed with a larger sample size. Test-retest should be reported to verify which post-processing pipeline is optimal for edited MRS, DW-MRS as well as conventional MRS.Conclusion
This study has examined different coil combination and spectral registration methods for DW-MRS post-processing. The tdCoilComb-svdSR pipeline achieved best post-processing of in-vivo DW-MRS data. The proposed SVD spectral registration method improved the spectral quality but needs some further verifications.Acknowledgements
No acknowledgement found.References
1. Simpson, R., Devenyi, G. A., Jezzard, P., Hennessy, T. J. & Near, J. Advanced processing and simulation of MRS data using the FID appliance ( FID‐A )—An open source, MATLAB ‐based toolkit. Magn. Reson. Med. 77, 23–33 (2017).
2. Edden, R. A. E., Puts, N. A. J., Harris, A. D., Barker, P. B. & Evans, C. J. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid–edited MR spectroscopy spectra. Journal of Magnetic Resonance Imaging 40, 1445–1452 (2014).
3. Oeltzschner, G. et al. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. Journal of Neuroscience Methods 343, 108827 (2020).
4. Brown, M. A. Time-domain combination of MR spectroscopy data acquired using phased-array coils. Magn Reson Med 52, 1207–1213 (2004).
5. Hall, E. L., Stephenson, M. C., Price, D. & Morris, P. G. Methodology for improved detection of low concentration metabolites in MRS: Optimised combination of signals from multi-element coil arrays. NeuroImage 86, 35–42 (2014).
6. Bydder, M., Hamilton, G., Yokoo, T. & Sirlin, C. B. Optimal phased-array combination for spectroscopy. Magnetic Resonance Imaging 26, 847–850 (2008).
7. Mikkelsen, M. et al. Correcting frequency and phase offsets in MRS data using robust spectral registration. NMR Biomed 33, e4368 (2020).
8. Rodgers, C. T. & Robson, M. D. Receive array magnetic resonance spectroscopy: Whitened singular value decomposition (WSVD) gives optimal Bayesian solution. Magn Reson Med 63, 881–891 (2010).
9. An, L., Willem van der Veen, J., Li, S., Thomasson, D. M. & Shen, J. Combination of multichannel single-voxel MRS signals using generalized least squares. J Magn Reson Imaging 37, 1445–1450 (2013).
10. Near, J. et al. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain: MRS Drift Correction Using Spectral Registration. Magn. Reson. Med. 73, 44–50 (2015).
11. Wilson, M. Robust retrospective frequency and phase correction for single-voxel MR spectroscopy: WILSON. Magn Reson Med 81, 2878–2886 (2019).
Figures