3938
Development of a 2D MRSI sequence with Chemical-Shift Selective Adiabatic Pulses (2𝜋-CSAP) using Pulseq at 7T1Department of High Field MR, Centre for Image Sciences, University of Medical Centre Utrecht, Utrecht, Netherlands, 2Institute for Diagnostic and Interventional Neuroradiology, Support Center for Advanced Neuroimaging (SCAN), University of Bern, Bern, Switzerland, 3Translational Imaging Center, sitem-insel AG, Bern, Switzerland, 4Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 5Lee Gil Ya Cancer & Diabetes Institute, Gachon University, Incheon, Korea, Republic of, 6Department of Health Sciences and Technology, GAIHST, Gachon University, Incheon, Korea, Republic of, 7Department of Radiology, Division of Medical Physics, University Medical Center Freiburg, Freiburg, Germany
Synopsis
Keywords: Data Acquisition, Spectroscopy
An MRSI sequence using a pair of chemically selective adiabatic 2𝜋 refocus pulses, referred to as 2𝜋-CSAP, was implemented using the open-source Pulseq framework. This sequence enables full coverage of the frequency spectrum of interest through a shift in carrier frequency, which has been a limitation in rapid MRSI sequences. In addition, this shifted frequency spectrum also minimizes signal contaminations from residual water signal after water suppression or unsuppressed water signal and strong lipid signals, possibly eliminating the need for additional water suppression pre-pulses. This base sequence can potentially be combined with accelerated acquisition techniques for better scan efficiency.Introduction
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive technique for measuring and visualizing metabolite level distribution. It can be used in the brain to provide information about tumour metabolism, neurodegenerative diseases, and metabolic disorders. However, residual water signal after water suppression and strong lipid signals in the brain and skull obscure the accurate quantification of metabolite signals. Current solutions need a relatively long repetition time (TR), which particularly limits their applicability to clinical settings, especially when a high resolution is required. At the high magnetic field, several accelerated MRSI methods have been proposed1, but MRI vendors do not provide such methods and implementing these in the vendor's proprietary sequence programming environment is tedious and complex. In this work, as a response to the 2023 ISMRM Challenge "Repeat it With Me: Reproducibility Team Challenge", we reproduce a 2D MRSI sequence using a pair of chemically selective adiabatic 2𝜋 refocusing pulses (called 2𝜋-CSAP2,3) at 7T using the open-source Pulseq framework4. This MRSI sequence could be combined with various fast readout strategies such as EPI used in a recently published SLOW-editing EPSI sequence3.Methods
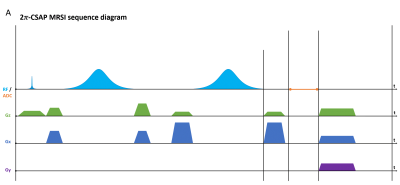
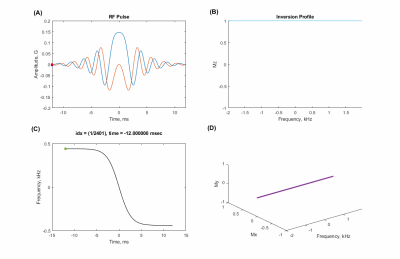
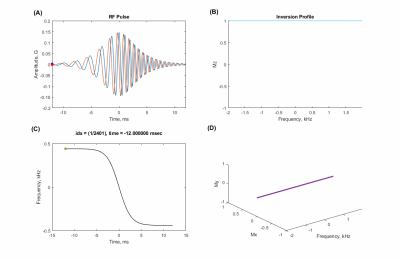
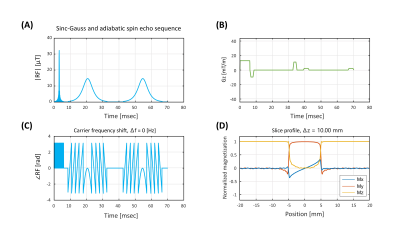
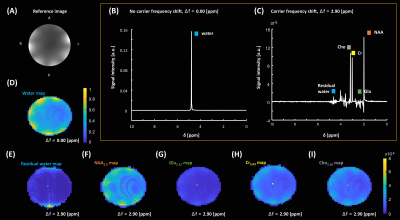
Sequence design, simulation, data acquisition, and reconstructionA 2𝜋-CSAP MRSI sequence (Fig. 1) was designed in the Pulseq framework. RF, GR, and ADC events were designed based on the parameters used in SLOW-editing3 full scheme 2. Bloch simulations were performed to validate the sequence timing and amplitudes of RF and gradient events before deploying into the scanner. An animation of magnetization evolution (Fig. 2, 3, and 4) during a 2𝜋-CSAP pulse was created with and without the carrier frequency shift (Δf = 0.00 [ppm] and 2.90 [ppm]). A 7T MRI scanner (Magnetom Terra, Siemens Healthineers, Erlangen, Germany) was used with a 1Tx/32Rx head coil (Nova Medical, USA) using the built-in gradient system (maximum gradient amplitude of 80 mT/m and maximum slew rate of 200 mT/m/msec). For the phantom experiment (BRAINO), imaging parameters were: FA = 65°, bandwidth = 5.5 kHz, and duration = 6 ms for a sinc-Gauss excitation pulse, and nominal FA = 550°, bandwidth = 0.88 kHz, and duration = 24 ms for a 2𝜋-CSAP pulse, FOV = 240 × 240 mm2, voxel size = 6 × 6 mm2, TR/TE = 1500/68 ms, spectral bandwidth = 5 kHz, number of sample points = 2048, readout oversampling factor = 2, number of averages = 1, and acquisition time = 40 min. An in-house developed script written in MATLAB was used for reconstruction. Raw data were exported as a TWIX format and converted to the ISMRMRD format5, a vendor-agnostic data format. A noise covariance matrix for pre-whitening was calculated from noise-only data. A spatial Fourier transform was performed, and the Roemer equal noise algorithm6,7 was used for channel combination using coil sensitivity maps8 estimated from the water reference. Zeroth-order phase correction was applied, and metabolite maps were generated by calculating the area of each metabolite peak.
Results and Discussion
When using the developed 2𝜋-CSAP sequence in the phantom (Fig. 5), selective excitation could be performed with minimal artifacts (e.g., residual water and lipid contamination) by shifting the carrier frequency. Water suppression pre-pulses were not included in this sequence. Using only 2 low-bandwidth adiabatic RF pulses reduces SAR and thus enables a relatively short TR. This is advantageous in acquiring a high resolution in vivo scan in a clinically acceptable scan time without compromising its ability to avoid the chemical shift displacement artifact (CSDA). A full bandwidth coverage is achieved in this sequence by setting the carrier frequency (Δf) to the center of the frequency band of interest (e.g., Δf = 2.9 [ppm], 1.6 - 4.2 [ppm]). The Pulseq framework based on MATLAB stores RF and gradient waveforms obtained from a Pulseq file, which enables quick Bloch simulations. This allows for a deep understanding of existing pulse sequences and possibly provides insights into a better pulse sequence design.Conclusion
We have successfully implemented a 2𝜋-CSAP sequence using an open-source sequence design tool. This pulse sequence written in Pulseq could be easily shared across different institutions having different scanners and scanner software versions. This sequence provides metabolic information and utilizes its spectral bandwidth by shifting the carrier frequency, and it avoids strong residual water signal, and lipid signal contamination. Moreover, the use of Pulseq can be easily extended to fast acquisition techniques such as EPI9, concentric ring10, and spiral11 k-space trajectory sequences.Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 813120.References
1. Bogner W, Otazo R, Henning A. Accelerated MR spectroscopic imaging—a review of current and emerging techniques. NMR Biomed. 2020;(March):1-32. doi:10.1002/nbm.4314
2. Conolly S, Nishimura D, Macovski A. A selective adiabatic spin-echo pulse. J Magn Reson. 1989;83(2):324-334. doi:10.1016/0022-2364(89)90194-7
3. Weng G, Radojewski P, Sheriff S, et al. SLOW: A novel spectral editing method for whole-brain MRSI at ultra high magnetic field. Magn Reson Med. 2022;88(1):53-70. doi:10.1002/mrm.29220
4. Layton KJ, Kroboth S, Jia F, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med. 2017;77(4):1544-1552. doi:10.1002/mrm.26235
5. Inati SJ, Naegele JD, Zwart NR, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn Reson Med. 2017;77(1):411-421. doi:10.1002/mrm.26089
6. Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16(2):192-225. doi:10.1002/mrm.1910160203
7. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952-962. doi:10.1002/(SICI)1522-2594(199911)42:5<952::AID-MRM16>3.0.CO;2-S
8. Brown MA. Time-domain combination of MR spectroscopy data acquired using phased-array coils. Magn Reson Med. 2004;52(5):1207-1213. doi:10.1002/mrm.20244
9. Posse S, Tedeschi G, Risinger R, Ogg R, Bihan D Le. High Speed 1H Spectroscopic Imaging in Human Brain by Echo Planar Spatial‐Spectral Encoding. Magn Reson Med. 1995;33(1):34-40. doi:10.1002/mrm.1910330106
10. Furuyama JK, Wilson NE, Thomas MA. Spectroscopic imaging using concentrically circular echo-planar trajectories in vivo. Magn Reson Med. 2012;67(6):1515-1522. doi:10.1002/mrm.23184
11. Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med. 1998;39(6):889-898. doi:10.1002/mrm.1910390606
Figures