3937
Compressed Sensing MPRAGE accelerates automated voxel placement for 1-H MRS of the human brain
Stefano Tambalo1, Sebastian Hübner1, Francesca Saviola1, Tobias Kober2,3,4, and Jorge Jovicich1
1University of Trento, Trento, Italy, 2Advanced Clinical Imaging Technology, Siemens Healthineers International AG, Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
1University of Trento, Trento, Italy, 2Advanced Clinical Imaging Technology, Siemens Healthineers International AG, Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Synopsis
Keywords: Data Processing, Spectroscopy
The automated voxel placement (AVP) framework has been proposed to optimize accuracy and reproducibility in single-voxel brain proton spectroscopy. AVP is based on the affine transformation of brain coordinates from standard to subject-specific space defined on a 3D T1-weighted structural scan. Here we evaluate the robustness of the AVP approach by evaluating the displacement of voxel center coordinates across 3D T1w MPRAGE sequence variants, including compressed sensing (CS) accelerated protocols. We show that AVP gave small but significantly higher voxel displacements for MP2RAGE and CS-MP2RAGE. There were no significant differences between multi-echo MPRAGE (6 min) and CS-MPRAGE (1 min).Introduction
Single-voxel proton magnetic resonance spectroscopy (MRS) results are crucially dependent on the accurate and reproducible positioning of the voxel that includes the target tissue of interest for metabolic evaluation [1]. Previous studies have shown that precision and reliability of brain MRS can be highly increased by using an automatic voxel placement (AVP) strategy [2]. This approach consists of acquiring the subject-specific T1-weighted (T1w) anatomical image, exporting it from the MR scanner to a computer that runs the AVP tool, setting the AVP to a predefined voxel coordinate in MNI space, computing the corresponding target voxel coordinates in subject space, and finally feeding these coordinates to the MR scanner console for the MRS acquisition. The AVP strategy has been demonstrated at 3T using a standard 3D T1w MPRAGE as input [2]. However, it remains unknown how robust the AVP is when different input T1w images are used, which is especially important with the availability of highly accelerated versions. In this study, for three different target MRS voxels, we study the variability of the AVP subject-specific coordinates as a function of MPRAGE sequence variants, both for standard (MPRAGE, multi-echo MPRAGE, MP2RAGE) and compressed sensing (CS) accelerated versions (CS-MP2RAGE and CS-MPRAGE).Methods
MRI acquisitionSix healthy adults (age=25.7±2 years) underwent 3T MRI (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) with a 64-channel head-neck coil. T1w images were acquired with three “standard” and three CS-accelerated 3D MPRAGE sequence variants using a research application sequence, all with 1mm isotropic voxels, same spatial coverage, active prescan normalize and no image filters: a) multi-echo MPRAGE (TR/TI=2530/1100, TE1-4=1.69/3.55/5.41/7.27ms, α=7°, GRAPPA=2, TA=6:03min) [3]; b) MP2RAGE (TR/TE=5000/2.98ms, α=4°/5°, GRAPPA=3, TA=8:52min) [4]; c) MPRAGE (TR/TE/TI=2310/3.48/1200ms, α=12°, GRAPPA=2, TA=5:32 min); d) CS-MP2RAGE (TR/TE=5000/2.88ms, α=4°/5°, undersampling factor 4.6, regularization factors 0.0006/0.0004, TA=3:40min) [5]; e) CS-MP2RAGE as in d), with online removal of background noise; f) CS-MPRAGE (TR/TE/TI=2300/2.88/900ms, α=9°, regularization=0.0006, undersampling factor 3.6, TA=2:04min) and g) CS-MPRAGE as in f) with undersampling factor 6.6, TA=1:14min [5]. More details on the CS reconstructions can be found in reference [5].
Single voxel placement for MRS
T1w images of each subject were given as input to an optimized version of the AVP tool [1] to enable faster coregistration of three hypothetical MRS voxels (templates) relevant for various diseases: right-hemispheric dorsolateral prefrontal cortex (rDLPFC, MNI=[36, 44, 20] [6], right-hemispheric basal ganglia (rBGAN, MNI=[20, 10, -2] [7]) and posterior cingulate cortex (PCC, MNI=[0, -53, 26] [8]). For each T1w sequence and voxel, the AVP algorithm translates standard MNI coordinates into subject space location.
Statistical analysis
The goal of the analysis was to quantify, for each MRS voxel location, the within-subject differences in the voxel position estimation as function of the various input T1w sequences. Relative displacements were measured computing the Euclidean distance between the affine transformation of voxel coordinates returned by AVP and the template voxel coordinates defined in standard space. The statistical analysis was performed as a two-way repeated measures ANOVA.
Results
Figure 1 shows, for a representative subject, the similarity of image contrast properties for the various sequences and acquisition times. As multi-echo MPRAGE we used the root mean squared calculated across echoes. As MP2RAGE we used the uniform image, which includes background noise, where the CS-MP2RAGE version provided a uniform and denoised image. Figure 2 shows the anatomical localization of the three template voxels in MNI space. Figure 3 shows the Euclidean distance differences (mm) for the three voxels across T1w sequences. Significant effect of sequence was found for the standard and CS-accelerated MPRAGE variants, regardless of the particular voxel location (p<0.01). Non significant effects were reported both for voxel location and sequence x voxel interaction (p=0.62; F(12, 105)=0.31, p=0.98, respectively).Discussion and conclusions
Our study shows that at the single-subject level, the AVP results depend on the details of the anatomical T1w sequence. With respect to the choice of the MPRAGE sequence, which was proposed in the original study [2], we found that MP2RAGE sequence variants, both standard and with CS acceleration, provide significantly different target voxel coordinates. The background noise from uniform MP2RAGE image, if not removed, potentially introduces a major source of error in estimating the affine transformations between spaces. A further step of denoising may diminish these differences, as suggested by the results of the uniform denoised version of the CS-MP2RAGE. There were no significant differences of voxel localization when using a fast CS-MPRAGE implementation of about 1 min acquisition time. Even though further acceleration may be possible combining CS-acceleration and reduced matrix size for AVP, higher resolution enables different applications - e.g. morphometry - that would otherwise require a separate acquisition. These results strongly support the use of fast CS-MPRAGE strategies for voxel positioning in brain MR spectroscopy.Acknowledgements
This work was supported by funding from the Municipality of the City of Rovereto (Trento), Italy, for the project “Advanced neuroimaging to study aging”
Disclosure
Tobias Kober is an employee of Siemens Healthineers International AG Switzerland, owns stocks of Siemens Healthineers and holds patents filed by Siemens Healthineers
References
- Ratai EM, Hancu I, Blezek DJ, Turk KW, Halpern E, et al. Automatic repositioning of MRSI voxels in longitudinal studies: impact on reproducibility of metabolite concentration measurements. J Magn Reson Imaging. 2008; 27(5):1188–1193.
- Woodcock EA, Arshad M, Khatib D, Stanley JA. Automated Voxel Placement: A Linux-based Suite of Tools for Accurate and Reliable Single Voxel Coregistration. J Neuroimaging Psychiatry Neurol. 2018;3(1):1-8.
- van der Kouwe AJW, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008 Apr 1;40(2):559-569.
- Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 2010 Jan 15;49(2):1271-81.
- Mussard E, Hilbert T, Forman C, Meuli R, Thiran JP, Kober T. Accelerated MP2RAGE imaging using Cartesian phyllotaxis readout and compressed sensing reconstruction. Magn Reson Med. 2020 Oct;84(4):1881-1894.
- Owen, A. M., McMillan, K. M., Laird, A. R., & Bullmore, E. N‐back working memory paradigm: A meta‐analysis of normative functional neuroimaging studies. Human brain mapping 2005, 25(1), 46-59.
- Elmaki, E. E., Gong, T., Nkonika, D. M., & Wang, G. Examining alterations in GABA concentrations in the basal ganglia of patients with Parkinson’s disease using MEGA-PRESS MRS. Japanese journal of radiology 2018, 36(3), 194-199
- Liu X, Duyn JH. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc Natl Acad Sci U S A. 2013 Mar 12;110(11):4392-7.
Figures
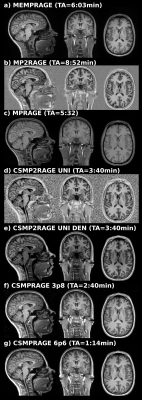
Figure 1: 3D MPRAGE sequence variants from a representative subject for visual qualitative comparison. Images shown with different brightness settings to better illustrate tissue contrast, which is well preserved for all sequences.
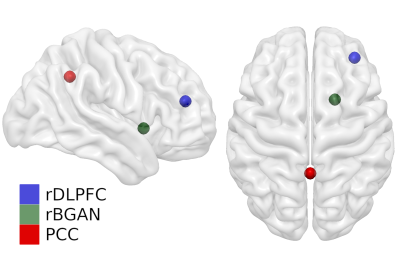
Figure 2. MRS voxel placement in MNI space for the three different template voxels. Abbreviations: rDLPFC, right-hemisphere dorsolateral prefrontal cortex; rBGAN, right-hemisphere basal ganglia and PCC, posterior cingulate cortex.
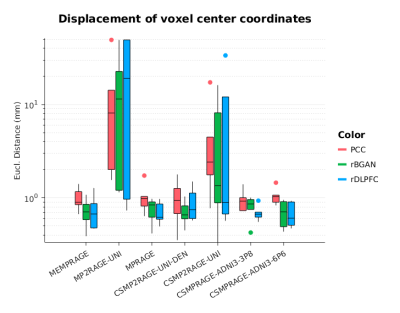
Figure 3. Euclidean distance differences (log scale, mm) calculated for each sequence variant and voxel location with respect to template voxel coordinates defined in MNI space.
DOI: https://doi.org/10.58530/2023/3937